[English] 日本語
 Yorodumi
Yorodumi- EMDB-16449: Cryo-EM structure of the human SIN3B full-length complex at 3.4 A... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human SIN3B full-length complex at 3.4 Angstrom resolution | |||||||||
 Map data Map data | SIN3B full-length complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Chromatin / Histone deacetylase / HDAC / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationautosome / positive regulation of male mating behavior / protein de-2-hydroxyisobutyrylase activity / protein lysine delactylase activity / negative regulation of dendritic spine development / p75NTR negatively regulates cell cycle via SC1 / epidermal cell differentiation / histone decrotonylase activity / fungiform papilla formation / NuRD complex ...autosome / positive regulation of male mating behavior / protein de-2-hydroxyisobutyrylase activity / protein lysine delactylase activity / negative regulation of dendritic spine development / p75NTR negatively regulates cell cycle via SC1 / epidermal cell differentiation / histone decrotonylase activity / fungiform papilla formation / NuRD complex / regulation of cell fate specification / behavioral response to ethanol / positive regulation of interleukin-1 production / negative regulation of transcription by competitive promoter binding / EGR2 and SOX10-mediated initiation of Schwann cell myelination / negative regulation of stem cell population maintenance / histone deacetylase activity, hydrolytic mechanism / ESC/E(Z) complex / histone deacetylase / regulation of stem cell differentiation / cellular response to dopamine / cardiac muscle hypertrophy / STAT3 nuclear events downstream of ALK signaling / XY body / regulation of double-strand break repair / protein lysine deacetylase activity / Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In linear amides / response to caffeine / histone deacetylase activity / embryonic digit morphogenesis / positive regulation of intracellular estrogen receptor signaling pathway / Notch-HLH transcription pathway / Y chromosome / X chromosome / histone deacetylase complex / eyelid development in camera-type eye / odontogenesis of dentin-containing tooth / Sin3-type complex / positive regulation of stem cell population maintenance / NuA4 histone acetyltransferase complex / dendrite development / response to amyloid-beta / positive regulation of oligodendrocyte differentiation / RNA Polymerase I Transcription Initiation / positive regulation of proteolysis / progesterone receptor signaling pathway / Regulation of MECP2 expression and activity / response to hyperoxia / hair follicle placode formation / NF-kappaB binding / FOXO-mediated transcription of oxidative stress, metabolic and neuronal genes / positive regulation of epithelial to mesenchymal transition / cellular response to transforming growth factor beta stimulus / Transcriptional regulation of brown and beige adipocyte differentiation by EBF2 / positive regulation of double-strand break repair via homologous recombination / MECP2 regulates neuronal receptors and channels / Regulation of TP53 Activity through Acetylation / cellular response to retinoic acid / heat shock protein binding / Regulation of lipid metabolism by PPARalpha / transcription repressor complex / phosphatidylinositol binding / negative regulation of cell migration / SUMOylation of chromatin organization proteins / Regulation of PTEN gene transcription / transcription corepressor binding / response to amphetamine / transcription coregulator binding / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / response to nicotine / Regulation of endogenous retroelements by KRAB-ZFP proteins / response to cocaine / Negative Regulation of CDH1 Gene Transcription / circadian regulation of gene expression / HDACs deacetylate histones / promoter-specific chromatin binding / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / negative regulation of transforming growth factor beta receptor signaling pathway / Cytoprotection by HMOX1 / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / double-strand break repair via homologous recombination / NoRC negatively regulates rRNA expression / NOTCH1 Intracellular Domain Regulates Transcription / Constitutive Signaling by NOTCH1 PEST Domain Mutants / Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants / histone deacetylase binding / cellular response to hydrogen peroxide / positive regulation of tumor necrosis factor production / transcription corepressor activity / heterochromatin formation / nucleosome / negative regulation of neuron projection development / cellular response to heat / HATs acetylate histones / Factors involved in megakaryocyte development and platelet production / chromatin organization / fibroblast proliferation / regulation of apoptotic process / histone binding / response to lipopolysaccharide Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Alfieri C / Wan SM / Muhammad R | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Mechanism of assembly, activation and lysine selection by the SIN3B histone deacetylase complex. Authors: Mandy S M Wan / Reyhan Muhammad / Marios G Koliopoulos / Theodoros I Roumeliotis / Jyoti S Choudhary / Claudio Alfieri /  Abstract: Lysine acetylation in histone tails is a key post-translational modification that controls transcription activation. Histone deacetylase complexes remove histone acetylation, thereby repressing ...Lysine acetylation in histone tails is a key post-translational modification that controls transcription activation. Histone deacetylase complexes remove histone acetylation, thereby repressing transcription and regulating the transcriptional output of each gene. Although these complexes are drug targets and crucial regulators of organismal physiology, their structure and mechanisms of action are largely unclear. Here, we present the structure of a complete human SIN3B histone deacetylase holo-complex with and without a substrate mimic. Remarkably, SIN3B encircles the deacetylase and contacts its allosteric basic patch thereby stimulating catalysis. A SIN3B loop inserts into the catalytic tunnel, rearranges to accommodate the acetyl-lysine moiety, and stabilises the substrate for specific deacetylation, which is guided by a substrate receptor subunit. Our findings provide a model of specificity for a main transcriptional regulator conserved from yeast to human and a resource of protein-protein interactions for future drug designs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16449.map.gz emd_16449.map.gz | 26.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16449-v30.xml emd-16449-v30.xml emd-16449.xml emd-16449.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16449.png emd_16449.png | 80.6 KB | ||
| Filedesc metadata |  emd-16449.cif.gz emd-16449.cif.gz | 8.1 KB | ||
| Others |  emd_16449_half_map_1.map.gz emd_16449_half_map_1.map.gz emd_16449_half_map_2.map.gz emd_16449_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16449 http://ftp.pdbj.org/pub/emdb/structures/EMD-16449 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16449 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16449 | HTTPS FTP |
-Related structure data
| Related structure data |  8c60MC  8bpaC  8bpbC  8bpcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16449.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16449.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SIN3B full-length complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.134 Å | ||||||||||||||||||||||||||||||||||||
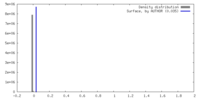
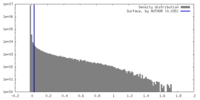
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: SIN3B full-length complex half map 1
| File | emd_16449_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SIN3B full-length complex half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
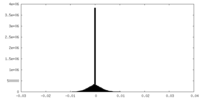
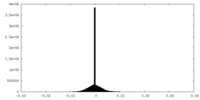
| Density Histograms |
-Half map: SIN3B full-length complex half map 2
| File | emd_16449_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SIN3B full-length complex half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
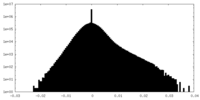
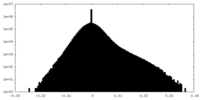
| Density Histograms |
- Sample components
Sample components
-Entire : Human SIN3B complex
| Entire | Name: Human SIN3B complex |
|---|---|
| Components |
|
-Supramolecule #1: Human SIN3B complex
| Supramolecule | Name: Human SIN3B complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 385 KDa |
-Macromolecule #1: Isoform 2 of Paired amphipathic helix protein Sin3b
| Macromolecule | Name: Isoform 2 of Paired amphipathic helix protein Sin3b / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 129.547133 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAHAGGGSGG SGAGGPAGRG LSGARWGRSG SAGHEKLPVH VEDALTYLDQ VKIRFGSDPA TYNGFLEIMK EFKSQSIDTP GVIRRVSQL FHEHPDLIVG FNAFLPLGYR IDIPKNGKLN IQSPLTSQEN SHNHGDGAED FKQQVPYKED KPQVPLESDS V EFNNAISY ...String: MAHAGGGSGG SGAGGPAGRG LSGARWGRSG SAGHEKLPVH VEDALTYLDQ VKIRFGSDPA TYNGFLEIMK EFKSQSIDTP GVIRRVSQL FHEHPDLIVG FNAFLPLGYR IDIPKNGKLN IQSPLTSQEN SHNHGDGAED FKQQVPYKED KPQVPLESDS V EFNNAISY VNKIKTRFLD HPEIYRSFLE ILHTYQKEQL NTRGRPFRGM SEEEVFTEVA NLFRGQEDLL SEFGQFLPEA KR SLFTGNG PCEMHSVQKN EHDKTPEHSR KRSRPSLLRP VSAPAKKKMK LRGTKDLSIA AVGKYGTLQE FSFFDKVRRV LKS QEVYEN FLRCIALFNQ ELVSGSELLQ LVSPFLGKFP ELFAQFKSFL GVKELSFAPP MSDRSGDGIS REIDYASCKR IGSS YRALP KTYQQPKCSG RTAICKEVLN DTWVSFPSWS EDSTFVSSKK TPYEEQLHRC EDERFELDVV LETNLATIRV LESVQ KKLS RMAPEDQEKF RLDDSLGGTS EVIQRRAIYR IYGDKAPEII ESLKKNPVTA VPVVLKRLKA KEEEWREAQQ GFNKIW REQ YEKAYLKSLD HQAVNFKQND TKALRSKSLL NEIESVYDEH QEQHSEGRSA PSSEPHLIFV YEDRQILEDA AALISYY VK RQPAIQKEDQ GTIHQLLHQF VPSLFFSQQL DLGASEESAD EDRDSPQGQT TDPSERKKPA PGPHSSPPEE KGAFGDAP A TEQPPLPPPA PHKPLDDVYS LFFANNNWYF FLRLHQTLCS RLLKIYRQAQ KQLLEYRTEK EREKLLCEGR REKGSDPAM ELRLKQPSEV ELEEYYPAFL DMVRSLLEGS IDPTQYEDTL REMFTIHAYV GFTMDKLVQN IARQLHHLVS DDVCLKVVEL YLNEKKRGA AGGNLSSRCV RAARETSYQW KAERCMADEN CFKVMFLQRK GQVIMTIELL DTEEAQTEDP VEVQHLARYV E QYVGTEGA SSSPTEGFLL KPVFLQRNLK KFRRRWQSEQ ARALRGEARS SWKRLVGVES ACDVDCRFKL STHKMVFIVN SE DYMYRRG TLCRAKQVQP LVLLRHHQHF EEWHSRWLED NVTVEAASLV QDWLMGEEDE DMVPCKTLCE TVHVHGLPVT RYR VQYSRR PASP UniProtKB: Paired amphipathic helix protein Sin3b |
-Macromolecule #2: Histone deacetylase 2
| Macromolecule | Name: Histone deacetylase 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: histone deacetylase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.443156 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAYSQGGGKK KVCYYYDGDI GNYYYGQGHP MKPHRIRMTH NLLLNYGLYR KMEIYRPHKA TAEEMTKYHS DEYIKFLRSI RPDNMSEYS KQMQRFNVGE DCPVFDGLFE FCQLSTGGSV AGAVKLNRQQ TDMAVNWAGG LHHAKKSEAS GFCYVNDIVL A ILELLKYH ...String: MAYSQGGGKK KVCYYYDGDI GNYYYGQGHP MKPHRIRMTH NLLLNYGLYR KMEIYRPHKA TAEEMTKYHS DEYIKFLRSI RPDNMSEYS KQMQRFNVGE DCPVFDGLFE FCQLSTGGSV AGAVKLNRQQ TDMAVNWAGG LHHAKKSEAS GFCYVNDIVL A ILELLKYH QRVLYIDIDI HHGDGVEEAF YTTDRVMTVS FHKYGEYFPG TGDLRDIGAG KGKYYAVNFP MRDGIDDESY GQ IFKPIIS KVMEMYQPSA VVLQCGADSL SGDRLGCFNL TVKGHAKCVE VVKTFNLPLL MLGGGGYTIR NVARCWTYET AVA LDCEIP NELPYNDYFE YFGPDFKLHI SPSNMTNQNT PEYMEKIKQR LFENLRMLPH APGVQMQAIP EDAVHEDSGD EDGE DPDKR ISIRASDKRI ACDEEFSDSE DEGEGGRRNV ADHKKGAKKA RIEEDKKETE DKKTDVKEED KSKDNSGEKT DTKGT KSEQ LSNP UniProtKB: Histone deacetylase 2 |
-Macromolecule #3: PHD finger protein 12
| Macromolecule | Name: PHD finger protein 12 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 109.841586 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MWEKMETKTI VYDLDTSGGL MEQIQALLAP PKTDEAEKRS RKPEKEPRRS GRATNHDSCD SCKEGGDLLC CDHCPAAFHL QCCNPPLSE EMLPPGEWMC HRCTVRRKKR EQKKELGHVN GLVDKSGKRT TSPSSDTDLL DRSASKTELK AIAHARILER R ASRPGTPT ...String: MWEKMETKTI VYDLDTSGGL MEQIQALLAP PKTDEAEKRS RKPEKEPRRS GRATNHDSCD SCKEGGDLLC CDHCPAAFHL QCCNPPLSE EMLPPGEWMC HRCTVRRKKR EQKKELGHVN GLVDKSGKRT TSPSSDTDLL DRSASKTELK AIAHARILER R ASRPGTPT SSASTETPTS EQNDVDEDII DVDEEPVAAE PDYVQPQLRR PFELLIAAAM ERNPTQFQLP NELTCTTALP GS SKRRRKE ETTGKNVKKT QHELDHNGLV PLPVKVCFTC NRSCRVAPLI QCDYCPLLFH MDCLEPPLTA MPLGRWMCPN HIE HVVLNQ KNMTLSNRCQ VFDRFQDTVS QHVVKVDFLN RIHKKHPPNR RVLQSVKRRS LKVPDAIKSQ YQFPPPLIAP AAIR DGELI CNGIPEESQM HLLNSEHLAT QAEQQEWLCS VVALQCSILK HLSAKQMPSH WDSEQTEKAD IKPVIVTDSS VTTSL QTAD KTPTPSHYPL SCPSGISTQN SLSCSPPHQS PALEDIGCSS CAEKSKKTPC GTANGPVNTE VKANGPHLYS SPTDST DPR RLPGANTPLP GLSHRQGWPR PLTPPAAGGL QNHTVGIIVK TENATGPSSC PQRSLVPVPS LPPSIPSSCA SIENTST LQ RKTVQSQIGP PLTDSRPLGS PPNATRVLTP PQAAGDGILA TTANQRFSSP APSSDGKVSP GTLSIGSALT VPSFPANS T AMVDLTNSLR AFMDVNGEIE INMLDEKLIK FLALQRIHQL FPSRVQPSPG SVGTHQLASG GHHIEVQRKE VQARAVFYP LLGLGGAVNM CYRTLYIGTG ADMDVCLTNY GHCNYVSGKH ACIFYDENTK HYELLNYSEH GTTVDNVLYS CDFSEKTPPT PPSSIVAKV QSVIRRRRHQ KQDEEPSEEA AMMSSQAQGP QRRPCNCKAS SSSLIGGSGA GWEGTALLHH GSYIKLGCLQ F VFSITEFA TKQPKGDASL LQDGVLAEKL SLKPHQGPVL RSNSVP UniProtKB: PHD finger protein 12 |
-Macromolecule #4: Mortality factor 4-like protein 1
| Macromolecule | Name: Mortality factor 4-like protein 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.540484 KDa |
| Recombinant expression | Organism:  Trichoplusia (butterflies/moths) Trichoplusia (butterflies/moths) |
| Sequence | String: MAPKQDPKPK FQEGERVLCF HGPLLYEAKC VKVAIKDKQV KYFIHYSGWN KKSAVRPRRS EKSLKTHEDI VALFPVPEGA PSVHHPLLT SSWDEWVPES RVLKYVDTNL QKQRELQKAN QEQYAEGKMR GAAPGKKTSG LQQKNVEVKT KKNKQKTPGN G DGGSTSET ...String: MAPKQDPKPK FQEGERVLCF HGPLLYEAKC VKVAIKDKQV KYFIHYSGWN KKSAVRPRRS EKSLKTHEDI VALFPVPEGA PSVHHPLLT SSWDEWVPES RVLKYVDTNL QKQRELQKAN QEQYAEGKMR GAAPGKKTSG LQQKNVEVKT KKNKQKTPGN G DGGSTSET PQPPRKKRAR VDPTVENEET FMNRVEVKVK IPEELKPWLV DDWDLITRQK QLFYLPAKKN VDSILEDYAN YK KSRGNTD NKEYAVNEVV AGIKEYFNVM LGTQLLYKFE RPQYAEILAD HPDAPMSQVY GAPHLLRLFV RIGAMLAYTP LDE KSLALL LNYLHDFLKY LAKNSATLFS ASDYEVAPPE YHRKAV UniProtKB: Mortality factor 4-like protein 1 |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 5 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)