[English] 日本語
 Yorodumi
Yorodumi- EMDB-16435: microtubule decorated with tubulin oligomers in presence of APC C... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
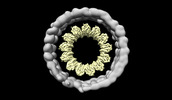
| Title | microtubule decorated with tubulin oligomers in presence of APC C-terminal domain. (here only the microtubule map section is represented) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cytoskeleton / microtubule / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytoplasmic microtubule / cellular response to interleukin-4 / structural constituent of cytoskeleton / microtubule cytoskeleton organization / neuron migration / mitotic cell cycle / double-stranded RNA binding / microtubule cytoskeleton / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule ...cytoplasmic microtubule / cellular response to interleukin-4 / structural constituent of cytoskeleton / microtubule cytoskeleton organization / neuron migration / mitotic cell cycle / double-stranded RNA binding / microtubule cytoskeleton / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / cilium / GTPase activity / ubiquitin protein ligase binding / GTP binding / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 5.3 Å | |||||||||
 Authors Authors | Serre L / Arnal I | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: J Cell Sci / Year: 2023 Journal: J Cell Sci / Year: 2023Title: The mitotic role of adenomatous polyposis coli requires its bilateral interaction with tubulin and microtubules. Authors: Laurence Serre / Julie Delaroche / Angélique Vinit / Guy Schoehn / Eric Denarier / Anne Fourest-Lieuvin / Isabelle Arnal /  Abstract: Adenomatous polyposis coli (APC) is a scaffold protein with tumour suppressor properties. Mutations causing the loss of its C-terminal domain (APC-C), which bears cytoskeleton-regulating sequences, ...Adenomatous polyposis coli (APC) is a scaffold protein with tumour suppressor properties. Mutations causing the loss of its C-terminal domain (APC-C), which bears cytoskeleton-regulating sequences, correlate with colorectal cancer. The cellular roles of APC in mitosis are widely studied, but the molecular mechanisms of its interaction with the cytoskeleton are poorly understood. Here, we investigated how APC-C regulates microtubule properties, and found that it promotes both microtubule growth and shrinkage. Strikingly, APC-C accumulates at shrinking microtubule extremities, a common characteristic of depolymerases. Cryo-electron microscopy revealed that APC-C adopts an extended conformation along the protofilament crest and showed the presence of ring-like tubulin oligomers around the microtubule wall, which required the presence of two APC-C sub-domains. A mutant of APC-C that was incapable of decorating microtubules with ring-like tubulin oligomers exhibited a reduced effect on microtubule dynamics. Finally, whereas native APC-C rescued defective chromosome alignment in metaphase cells silenced for APC, the ring-incompetent mutant failed to correct mitotic defects. Thus, the bilateral interaction of APC-C with tubulin and microtubules likely contributes to its mitotic functions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16435.map.gz emd_16435.map.gz | 114.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16435-v30.xml emd-16435-v30.xml emd-16435.xml emd-16435.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16435.png emd_16435.png | 48.2 KB | ||
| Masks |  emd_16435_msk_1.map emd_16435_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16435.cif.gz emd-16435.cif.gz | 6.7 KB | ||
| Others |  emd_16435_half_map_1.map.gz emd_16435_half_map_1.map.gz emd_16435_half_map_2.map.gz emd_16435_half_map_2.map.gz | 97.5 MB 97.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16435 http://ftp.pdbj.org/pub/emdb/structures/EMD-16435 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16435 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16435 | HTTPS FTP |
-Related structure data
| Related structure data |  8c5cMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16435.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16435.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.96 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16435_msk_1.map emd_16435_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16435_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16435_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : microtubule
| Entire | Name: microtubule |
|---|---|
| Components |
|
-Supramolecule #1: microtubule
| Supramolecule | Name: microtubule / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Tubulin beta chain
| Macromolecule | Name: Tubulin beta chain / type: protein_or_peptide / ID: 1 / Number of copies: 26 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.019922 KDa |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEATGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEATGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS VMPSPKVSDT VVEPYNATLS VHQLVENTDE TYCIDNEALY DICFRTLKLT TPTYGDLNHL VSATMSGVTT CL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTSRG SQQYRALTVP ELTQQMFDSK NMMAACDPRH GRYLTVAAIF RGR MSMKEV DEQMLNVQNK NSSYFVEWIP NNVKTAVCDI PPRGLKMSAT FIGNSTAIQE LFKRISEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY QDATADEQGE GHEDEEKDEG G UniProtKB: Tubulin beta 4B class IVb |
-Macromolecule #2: Tubulin alpha-1B chain
| Macromolecule | Name: Tubulin alpha-1B chain / type: protein_or_peptide / ID: 2 / Number of copies: 26 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.204445 KDa |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE FSIYPAPQVS TAVVEPYNSI LTTHTTLEHS DCAFMVDNEA IYDICRRNLD IERPTYTNLN RLISQIVSSI TA SLRFDGA LNVDLTEFQT NLVPYPRIHF PLATYAPVIS AEKAYHEQLS VAEITNACFE PANQMVKCDP RHGKYMACCL LYR GDVVPK DVNAAIATIK TKRSIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVQRAVC MLSNTTAIAE AWARLDHKFD LMYA KRAFV HWYVGEGMEE GEFSEAREDM AALEKDYEEV GVDSVEGEGE EEGEEY UniProtKB: Tubulin alpha-1B chain |
-Macromolecule #3: TAXOL
| Macromolecule | Name: TAXOL / type: ligand / ID: 3 / Number of copies: 13 / Formula: TA1 |
|---|---|
| Molecular weight | Theoretical: 853.906 Da |
| Chemical component information |  ChemComp-TA1: |
-Macromolecule #4: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 13 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 13 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 13 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.75 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 37 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 9.54 Å Applied symmetry - Helical parameters - Δ&Phi: -27.68 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 5.3 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Details: resolution deduced from unmasked FSC is 7.1 A / Number images used: 11670 |
|---|---|
| CTF correction | Software - Name: CTFFIND / Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
| Startup model | Type of model: OTHER / Details: MAP from MIRP pipeline (cook et al. 2020) |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)