+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
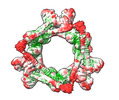
| Title | HSF2BP-BRCA2 ring-shaped complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Meiosis / Cancer Antigen / DNA repair / Fertility / Armadillo domain / Intrinsically Disordered Protein / RECOMBINATION | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
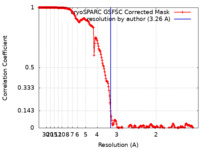
| Method | single particle reconstruction / cryo EM / Resolution: 3.26 Å | |||||||||
 Authors Authors | Zinn-Justin S / Ghouil R / Miron S / Legrand P / Ouldali M / Winter JM / Ropars V / Arteni AA | |||||||||
| Funding support | European Union,  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination. Authors: Rania Ghouil / Simona Miron / Koichi Sato / Dejan Ristic / Sari E van Rossum-Fikkert / Pierre Legrand / Malika Ouldali / Jean-Marie Winter / Virginie Ropars / Gabriel David / Ana-Andreea ...Authors: Rania Ghouil / Simona Miron / Koichi Sato / Dejan Ristic / Sari E van Rossum-Fikkert / Pierre Legrand / Malika Ouldali / Jean-Marie Winter / Virginie Ropars / Gabriel David / Ana-Andreea Arteni / Claire Wyman / Puck Knipscheer / Roland Kanaar / Alex N Zelensky / Sophie Zinn-Justin /   Abstract: In meiotic homologous recombination (HR), BRCA2 facilitates loading of the recombinases RAD51 and DMC1 at the sites of double-strand breaks (DSBs). The HSF2BP-BRME1 complex interacts with BRCA2. Its ...In meiotic homologous recombination (HR), BRCA2 facilitates loading of the recombinases RAD51 and DMC1 at the sites of double-strand breaks (DSBs). The HSF2BP-BRME1 complex interacts with BRCA2. Its absence causes a severe reduction in recombinase loading at meiotic DSB. We previously showed that, in somatic cancer cells ectopically producing HSF2BP, DNA damage can trigger HSF2BP-dependent degradation of BRCA2, which prevents HR. Here, we report that, upon binding to BRCA2, HSF2BP forms octameric rings that are able to interlock into a large ring-shaped 24-mer. Addition of BRME1 leads to dissociation of both of these ring structures and cancels the disruptive effect of HSF2BP on cancer cell resistance to DNA damage. It also prevents BRCA2 degradation during interstrand DNA crosslink repair in egg extracts. We propose that, during meiosis, the control of HSF2BPBRCA2 oligomerization by BRME1 ensures timely assembly of the ring complex that concentrates BRCA2 and controls its turnover, thus promoting HR. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16432.map.gz emd_16432.map.gz | 404.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16432-v30.xml emd-16432-v30.xml emd-16432.xml emd-16432.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16432_fsc.xml emd_16432_fsc.xml | 19.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16432.png emd_16432.png | 121.7 KB | ||
| Filedesc metadata |  emd-16432.cif.gz emd-16432.cif.gz | 5.7 KB | ||
| Others |  emd_16432_half_map_1.map.gz emd_16432_half_map_1.map.gz emd_16432_half_map_2.map.gz emd_16432_half_map_2.map.gz | 763.6 MB 763.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16432 http://ftp.pdbj.org/pub/emdb/structures/EMD-16432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16432 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16432.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16432.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.73 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16432_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
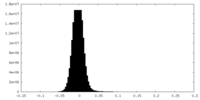
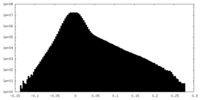
| Density Histograms |
-Half map: #2
| File | emd_16432_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
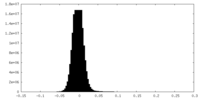
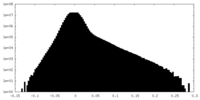
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between HSF2BP and the BRCA2 fragment N2291-Q2342
| Entire | Name: Complex between HSF2BP and the BRCA2 fragment N2291-Q2342 |
|---|---|
| Components |
|
-Supramolecule #1: Complex between HSF2BP and the BRCA2 fragment N2291-Q2342
| Supramolecule | Name: Complex between HSF2BP and the BRCA2 fragment N2291-Q2342 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The complex contains 24 HSF2BP molecules and 12 BRCA2 peptides |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 900 kDa/nm |
-Macromolecule #1: HSF2BP
| Macromolecule | Name: HSF2BP / type: protein_or_peptide / ID: 1 / Details: Full-length human HSF2BP / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MGEAGAAEEA CRHMGTKEEF VKVRKKDLER LTTEVMQIRD FLPRILNGEV LESFQKLKIV EKNLERKEQ ELEQLKMDCE HFKARLETVQ ADNIREKKEK LALRQQLNEA KQQLLQQAEY C TEMGAAAC TLLWGVSSSE EVVKAILGGD KALKFFSITG QTMESFVKSL ...String: MGEAGAAEEA CRHMGTKEEF VKVRKKDLER LTTEVMQIRD FLPRILNGEV LESFQKLKIV EKNLERKEQ ELEQLKMDCE HFKARLETVQ ADNIREKKEK LALRQQLNEA KQQLLQQAEY C TEMGAAAC TLLWGVSSSE EVVKAILGGD KALKFFSITG QTMESFVKSL DGDVQELDSD ES QFVFALA GIVTNVAAIA CGREFLVNSS RVLLDTILQL LGDLKPGQCT KLKVLMLMSL YNV SINLKG LKYISESPGF IPLLWWLLSD PDAEVCLHVL RLVQSVVLEP EVFSKSASEF RSSL PLQRI LAMSKSRNPR LQTAAQELLE DLRTLEHNV |
-Macromolecule #2: BRCA2
| Macromolecule | Name: BRCA2 / type: protein_or_peptide / ID: 2 Details: BRCA2 from N2291 to Q2342 with a C-terminal TEV site (was produced fused to GB1-6His) Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: NEFDRIIENQ EKSLKASKST PDGTIKDRRL FMHHVSLEPI TTVPFRTTKE RQENLYFQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 300 | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | Obtained after gel filtration on the complex |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Number real images: 9531 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)