[English] 日本語
 Yorodumi
Yorodumi- EMDB-16370: Structure of CUL2-KLHDC2 E3 ligase autoinhibited by C-degron mimicry -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of CUL2-KLHDC2 E3 ligase autoinhibited by C-degron mimicry | |||||||||
 Map data Map data | relion postprocess | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cullin-RING E3 ubiquitin ligase / LIGASE / ubiquitin / KLHDC2 / C-degron / CUL2 | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
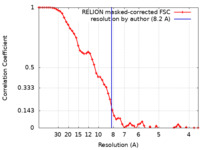
| Method | single particle reconstruction / cryo EM / Resolution: 8.2 Å | |||||||||
 Authors Authors | Scott DC / King M / Baek K / Schulman BA | |||||||||
| Funding support |  Germany, European Union, 2 items Germany, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: E3 ligase autoinhibition by C-degron mimicry maintains C-degron substrate fidelity. Authors: Daniel C Scott / Moeko T King / Kheewoong Baek / Clifford T Gee / Ravi Kalathur / Jerry Li / Nicholas Purser / Amanda Nourse / Sergio C Chai / Sivaraja Vaithiyalingam / Taosheng Chen / ...Authors: Daniel C Scott / Moeko T King / Kheewoong Baek / Clifford T Gee / Ravi Kalathur / Jerry Li / Nicholas Purser / Amanda Nourse / Sergio C Chai / Sivaraja Vaithiyalingam / Taosheng Chen / Richard E Lee / Stephen J Elledge / Gary Kleiger / Brenda A Schulman /   Abstract: E3 ligase recruitment of proteins containing terminal destabilizing motifs (degrons) is emerging as a major form of regulation. How those E3s discriminate bona fide substrates from other proteins ...E3 ligase recruitment of proteins containing terminal destabilizing motifs (degrons) is emerging as a major form of regulation. How those E3s discriminate bona fide substrates from other proteins with terminal degron-like sequences remains unclear. Here, we report that human KLHDC2, a CRL2 substrate receptor targeting C-terminal Gly-Gly degrons, is regulated through interconversion between two assemblies. In the self-inactivated homotetramer, KLHDC2's C-terminal Gly-Ser motif mimics a degron and engages the substrate-binding domain of another protomer. True substrates capture the monomeric CRL2, driving E3 activation by neddylation and subsequent substrate ubiquitylation. Non-substrates such as NEDD8 bind KLHDC2 with high affinity, but its slow on rate prevents productive association with CRL2. Without substrate, neddylated CRL2 assemblies are deactivated via distinct mechanisms: the monomer by deneddylation and the tetramer by auto-ubiquitylation. Thus, substrate specificity is amplified by KLHDC2 self-assembly acting like a molecular timer, where only bona fide substrates may bind before E3 ligase inactivation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16370.map.gz emd_16370.map.gz | 2.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16370-v30.xml emd-16370-v30.xml emd-16370.xml emd-16370.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16370_fsc.xml emd_16370_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_16370.png emd_16370.png | 141.8 KB | ||
| Masks |  emd_16370_msk_1.map emd_16370_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16370.cif.gz emd-16370.cif.gz | 4 KB | ||
| Others |  emd_16370_additional_1.map.gz emd_16370_additional_1.map.gz emd_16370_half_map_1.map.gz emd_16370_half_map_1.map.gz emd_16370_half_map_2.map.gz emd_16370_half_map_2.map.gz | 11.9 MB 11.9 MB 11.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16370 http://ftp.pdbj.org/pub/emdb/structures/EMD-16370 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16370 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16370 | HTTPS FTP |
-Validation report
| Summary document |  emd_16370_validation.pdf.gz emd_16370_validation.pdf.gz | 754.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16370_full_validation.pdf.gz emd_16370_full_validation.pdf.gz | 754.3 KB | Display | |
| Data in XML |  emd_16370_validation.xml.gz emd_16370_validation.xml.gz | 11.3 KB | Display | |
| Data in CIF |  emd_16370_validation.cif.gz emd_16370_validation.cif.gz | 15.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16370 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16370 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16370 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16370 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16370.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16370.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | relion postprocess | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.885 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16370_msk_1.map emd_16370_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3d refinement
| File | emd_16370_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3d refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfmap1
| File | emd_16370_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfmap2
| File | emd_16370_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of CUL2-KLHDC2 E3 ligase autoinhibited by C-degron mimicry
| Entire | Name: Structure of CUL2-KLHDC2 E3 ligase autoinhibited by C-degron mimicry |
|---|---|
| Components |
|
-Supramolecule #1: Structure of CUL2-KLHDC2 E3 ligase autoinhibited by C-degron mimicry
| Supramolecule | Name: Structure of CUL2-KLHDC2 E3 ligase autoinhibited by C-degron mimicry type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)