[English] 日本語
 Yorodumi
Yorodumi- EMDB-16259: Negative stain EM structure of Toxoplasma gondii glideosome-assoc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain EM structure of Toxoplasma gondii glideosome-associated connector subdomain coil 3 | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | actin-binding protein / armadillo repeat / solenoid / pleckstrin-homology domain / PROTEIN BINDING | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
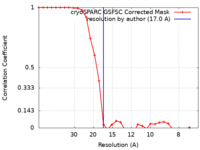
| Method | single particle reconstruction / negative staining / Resolution: 17.0 Å | ||||||||||||||||||||||||
 Authors Authors | Chen Q / Rosenthal P | ||||||||||||||||||||||||
| Funding support |  Finland, Finland,  Norway, Norway,  United Kingdom, 7 items United Kingdom, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of Toxoplasma gondii glideosome-associated connector suggests a role as an elastic element to assist myosin A in generating force for gliding motility Authors: Hung YF / Chen Q / Pires I / Rosenthal P / Kursula I | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16259.map.gz emd_16259.map.gz | 986.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16259-v30.xml emd-16259-v30.xml emd-16259.xml emd-16259.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16259_fsc.xml emd_16259_fsc.xml | 2.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_16259.png emd_16259.png | 15.3 KB | ||
| Filedesc metadata |  emd-16259.cif.gz emd-16259.cif.gz | 4.9 KB | ||
| Others |  emd_16259_half_map_1.map.gz emd_16259_half_map_1.map.gz emd_16259_half_map_2.map.gz emd_16259_half_map_2.map.gz | 1.8 MB 1.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16259 http://ftp.pdbj.org/pub/emdb/structures/EMD-16259 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16259 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16259 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16259.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16259.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.3 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16259_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16259_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Toxoplasma gondii glideosome-associated connector (coil 3)
| Entire | Name: Toxoplasma gondii glideosome-associated connector (coil 3) |
|---|---|
| Components |
|
-Supramolecule #1: Toxoplasma gondii glideosome-associated connector (coil 3)
| Supramolecule | Name: Toxoplasma gondii glideosome-associated connector (coil 3) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Toxoplasma gondii glideosome-associated connector (coil 3)
| Macromolecule | Name: Toxoplasma gondii glideosome-associated connector (coil 3) type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: QPREKTCKDV YDLLNSRVQ QGLSVAISEV AILQEEVEFL VSQ(MSE)G(MSE)YNQE QLDHQTA(MSE)GA DHQYGN(MSE)AFE LLAATSANVK LLQA NEFSK (MSE)ELALIKGQA DPEIVLYAVK ALTAFCKFPP AAQDTARIQG CPALVTEACS KINKSGLPNE ...String: QPREKTCKDV YDLLNSRVQ QGLSVAISEV AILQEEVEFL VSQ(MSE)G(MSE)YNQE QLDHQTA(MSE)GA DHQYGN(MSE)AFE LLAATSANVK LLQA NEFSK (MSE)ELALIKGQA DPEIVLYAVK ALTAFCKFPP AAQDTARIQG CPALVTEACS KINKSGLPNE RKEEHLCARY F LVERTAIN RNLYNKTPI(MSE) TELINSWNDY DKGAYTTTLL RFVFRA(MSE)RRV VSDAHVEELL KANVLQRLIG IISD VNAD(MSE) ALLPDVLFLL GSLAVVPEIK TKIGELNGIA ACTDLLQRAL PKPNTAPVVT NVCLAFANIC IGHKKNTEIF S KLGGPALN VKVLNDRGHE YDVCNAASVL LCNLLYKNES (MSE)KKLLGTNGA PAALVKGLSN YDGSEEKTAI RCLESVFK A ISNLSLYTPN IQPFLDAGIE NAYSTWLSNL SETFPDAQLE TGCRTLVNLV (MSE)ENEENN(MSE)RK FGVCLLPC (MSE) AVAKQGRTDT KALLLLLDIE ASLCRLKENA EAFAANGGIE TTIRLIHQFD YDVGLLTLGI HLLGIQSAVK DSIQR (MSE)(MSE)DA DVFSILVGCV EVDAEGNEVT DLVVGGLRCT RRIVRSEELA FEYCNAGGIA TIANVICKSI NQP(MSE) V(MSE)LEA CRVLLGLLFY TTRSQADRQA AVEALHAQCQ QRAEQ(MSE)HAQA QADYEAGVVS EPPPEE(MSE)EVP EPD PDELAN AAYGGWYQ(MSE)G (MSE)DEV(MSE)IDAIL QAVCACAAVE AHAKQLRLQR VCLGLAAYFA SEQ(MSE)GTSS L VGSGIEQVLT QI(MSE)TNFAGEG TT(MSE)QLSCVII NSIA(MSE)TSGD(MSE) YEEIKTSALL SALKTSVGK (MSE)ATKKPEEKA LKETCAATLE AAS |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Staining | Type: NEGATIVE / Material: sodium silicotungstate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (2k x 2k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)