[English] 日本語
 Yorodumi
Yorodumi- EMDB-16257: Cryo-EM structure of Toxoplasma gondii glideosome-associated connector -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Toxoplasma gondii glideosome-associated connector | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | actin-binding protein / armadillo repeat / solenoid / pleckstrin-homology domain / PROTEIN BINDING | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
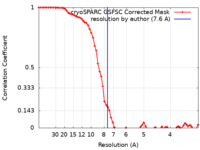
| Method | single particle reconstruction / cryo EM / Resolution: 7.6 Å | ||||||||||||||||||||||||
 Authors Authors | Chen Q / Rosenthal P | ||||||||||||||||||||||||
| Funding support |  Finland, Finland,  Norway, Norway,  United Kingdom, 7 items United Kingdom, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of Toxoplasma gondii glideosome-associated connector suggests a role as an elastic element to assist myosin A in generating force for gliding motility Authors: Hung YF / Chen Q / Pires I / Rosenthal P / Kursula I | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16257.map.gz emd_16257.map.gz | 28.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16257-v30.xml emd-16257-v30.xml emd-16257.xml emd-16257.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16257_fsc.xml emd_16257_fsc.xml | 6.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_16257.png emd_16257.png | 36.2 KB | ||
| Masks |  emd_16257_msk_1.map emd_16257_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16257.cif.gz emd-16257.cif.gz | 6.3 KB | ||
| Others |  emd_16257_half_map_1.map.gz emd_16257_half_map_1.map.gz emd_16257_half_map_2.map.gz emd_16257_half_map_2.map.gz | 28.3 MB 28.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16257 http://ftp.pdbj.org/pub/emdb/structures/EMD-16257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16257 | HTTPS FTP |
-Validation report
| Summary document |  emd_16257_validation.pdf.gz emd_16257_validation.pdf.gz | 658.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16257_full_validation.pdf.gz emd_16257_full_validation.pdf.gz | 657.9 KB | Display | |
| Data in XML |  emd_16257_validation.xml.gz emd_16257_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_16257_validation.cif.gz emd_16257_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16257 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16257 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16257 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16257 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16257.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16257.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.61 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
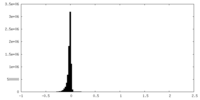
-Mask #1
| File |  emd_16257_msk_1.map emd_16257_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
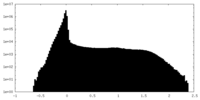
-Half map: #1
| File | emd_16257_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16257_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : toxoplasma gondii glideosome-associated connector
| Entire | Name: toxoplasma gondii glideosome-associated connector |
|---|---|
| Components |
|
-Supramolecule #1: toxoplasma gondii glideosome-associated connector
| Supramolecule | Name: toxoplasma gondii glideosome-associated connector / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: toxoplasma gondii glideosome-associated connector
| Macromolecule | Name: toxoplasma gondii glideosome-associated connector / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: G(MSE)AIGKVKRN PDAGVATAVQ SVIQHQTFKR (MSE)LLFGLRSLA DFCSPSNQLY QENALDALDR GVLSAIQTAV TT FSDDDDL (MSE)LCASRVLWA (MSE)SVAIKEE(MSE)D PAHIARVHSE GSPVIVAVVN SSPTDPQTIE DS(MSE)NFVD NL KRAGAPVDGA ...String: G(MSE)AIGKVKRN PDAGVATAVQ SVIQHQTFKR (MSE)LLFGLRSLA DFCSPSNQLY QENALDALDR GVLSAIQTAV TT FSDDDDL (MSE)LCASRVLWA (MSE)SVAIKEE(MSE)D PAHIARVHSE GSPVIVAVVN SSPTDPQTIE DS(MSE)NFVD NL KRAGAPVDGA SLAGG(MSE)LSIF TKTALD(MSE)KTA KRVTAALAIA AETAEGSTAL YNAGGTSVLL TYCLDQGDLS DAGVE(MSE)VEG AFDTVRY(MSE)AG YQCTDATTLP QCIAL(MSE)DKYR GRKSASAKGS SALAA(MSE)IGPE QLQKC LNTL KTAEAGSAEY DEALVTLGS(MSE) SYISSFTDEI VRAGGVPLLI ELINSGLPQ(MSE) EGNPEKIAS(MSE) ISGA AK(MSE)LA RIASNPVNVD AIVQAGGVAT LCTAVSYCTE S(MSE)EALGALC(MSE) ALVPLASRES LAHEIVQYQT FAT VLPILY QNVESPEIAA LA(MSE)ELVATGS QHEEIQEH(MSE)L QNQAAEICSL CCQYHTADAS YQQHAISALN RLVPRL TTL HGVSEYGGIQ GVIASLNANV NNEQVALLAV QLLDNFSEVS DAKTY(MSE)SDGT CVDAVLAA(MSE)L EHEGNDLLI SAGVHCLARI ATEDDCARHL NVLDTAIQTA RGNPDGVYRV LAAISGLSRV PSLRQIFEEK NASDTILAGI SSWIECSRFE GQNRIIKAA LKTVKN(MSE)KIS GDGDLTSCFA A(MSE)CDVACLPQ VKRVVELEEP DNNILVADTA AFRDLAAT(MSE) RITGAENLER CIESVLRV(MSE)R KYPDSRRAQL NCLETLNYLA QCDGGEGVAI LSRTGGLNAV VQYLTRAP(MSE)Y LD AQIAGFT VLATSAKIDS NVGETLRKCN CLQALKVA(MSE)R THAKSKELKR TIAPLVALL(MSE) PTDALETEIQ ELLNE CASA CEKNNFPHLH ENLAALNELL ISSEGAKIAA RLGIGAH(MSE)CK YQEYISAHEQ DALAVTDYDI LGKDLFDATV SE CAHA(MSE)EQ VASTRSGRNA LIKAGNVATL ISLYESLKAP QSQYSEEAAI HCLEALRILL KSDKRSAELA FERNFVSTL CVGIDSFPHS APVLGATCAC LAA(MSE)ATTPER VQ(MSE)LTAQPAF ESLLQKLVFV IQNDPSKDNK LVA(MSE)RALQ E LVEITNDAT(MSE) ANKIAEAGAV TALFRIIDEY GDDEQLTVQA AEVLALLGAF EDLRRFYDND VRFPAQVLTA ALTKQ KNNE TAVVHLLDVL NKLATSEDRA VLRELGV(MSE)EQ VADA(MSE)RVHSE SEAVTRLGGE LFAK(MSE)GADEQ IKSL (MSE)LQII ETVESGAEDT AQTVDILCGR LAVFLAAPLE DPRDALQHTE KCLGSLVATL QTYPGSERLE GNVALVCRRL C DRCFDDAD DPYGAWAVAA SG(MSE)LAQFAG(MSE) VAGETVLANK KFLGPAYRTF TACCANAYC(MSE) PT(MSE)VEV APS FLPQTYTLLE (MSE)HKNDAETVA RVLEFLRYFA EDPTACGLIV QN(MSE)SGSSGDV VALTVLL(MSE)QQ HQNND AVVC AG(MSE)EFLGALA YTLSQAGYEP LPTLADGSVL RDCDAL(MSE)GSN SSSARQLAH(MSE) H(MSE)IEK(MSE) LLS KAYNDALIQE QALKKLT(MSE)SL KAEDDKKRFS DEERAGLYAA (MSE)ACVLLAAGG AGLTGE(MSE)EKF NGFEV VLQA IEEFGENPTV IKEVNRALQG LS(MSE)ADVN(MSE)TA RTVKEAVPKL CTEATTAIQT DAECADTFCD L(MSE)LQ LVSQE GNGRQLLQVY GLEETLQGVE NLAAYYGEDF GTQLSEKVA(MSE) IRQA(MSE)EDDQP REKTCKDVYD LLNSRVQ QG LSVAISEVAI LQEEVEFLVS Q(MSE)G(MSE)YNQEQL DHQTA(MSE)GADH QYGN(MSE)AFELL AATSANVKLL QA NEFSK(MSE)E LALIKGQADP EIVLYAVKAL TAFCKFPPAA QDTARIQGCP ALVTEACSKI NKSGLPNERK EEHLCARYF LVERTAINRN LYNKTPI(MSE)TE LINSWNDYDK GAYTTTLLRF VFRA(MSE)RRVVS DAHVEELLKA NVLQRLIGII SD VNAD(MSE)AL LPDVLFLLGS LAVVPEIKTK IGELNGIAAC TDLLQRALPK PNTAPVVTNV CLAFANICIG HKKNTEIFS KLGGPALNVK VLNDRGHEYD VCNAASVLLC NLLYKNES(MSE)K KLLGTNGAPA ALVKGLSNYD GSEEKTAIRC LESVFK AIS NLSLYTPNIQ PFLDAGIENA YSTWLSNLSE TFPDAQLETG CRTLVNLV(MSE)E NEENN(MSE)RKFG VCLLPC (MSE)AV AKQGRTDTKA LLLLLDIEAS LCRLKENAEA FAANGGIETT IRLIHQFDYD VGLLTLGIHL LGIQSAVKDS IQR (MSE)(MSE)DADV FSILVGCVEV DAEGNEVTDL VVGGLRCTRR IVRSEELAFE YCNAGGIATI ANVICKSINQ P(MSE) V(MSE)LEACR VLLGLLFYTT RSQADRQAAV EALHAQCQQR AEQ(MSE)HAQAQA DYEAGVVSEP PPEE(MSE)EVPEP D PDELANAA YGGWYQ(MSE)G(MSE)D EV(MSE)IDAILQA VCACAAVEAH AKQLRLQRVC LGLAAYFASE Q(MSE)GTSS LVG SGIEQVLTQI (MSE)TNFAGEGTT (MSE)QLSCVIINS IA(MSE)TSGD(MSE)YE EIKTSALLSA LKTSVGK (MSE)A TKKPEEKALK ETCAATLEAA SSGEDPFDAF SKTVTELDFK FTEWNVDPYP NGVHDLPSNV KEALRKGGKL KVFL PEKEK EEIRWRSSQD LNVFEWC(MSE)GN DQDYNNRIPI VRIRNVAKGL VHPALKAAAK KEPRKVAAKF T(MSE)CLFGP PN DDFPEGVELP (MSE)VAKSQKERD AFVE(MSE)(MSE)VQWR DAATYNFHHH HHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)