+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the BUS complex - domain IV lattice | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DnaA / cryo-EM / BUS complex / replication initiation / REPLICATION | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.4 Å | |||||||||
 Authors Authors | Pelliciari S / Bodet-Lefevre S / Murray H / Ilangovan A | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: The bacterial replication origin BUS promotes nucleobase capture. Authors: Simone Pelliciari / Salomé Bodet-Lefèvre / Stepan Fenyk / Daniel Stevens / Charles Winterhalter / Frederic D Schramm / Sara Pintar / Daniel R Burnham / George Merces / Tomas T Richardson / ...Authors: Simone Pelliciari / Salomé Bodet-Lefèvre / Stepan Fenyk / Daniel Stevens / Charles Winterhalter / Frederic D Schramm / Sara Pintar / Daniel R Burnham / George Merces / Tomas T Richardson / Yumiko Tashiro / Julia Hubbard / Hasan Yardimci / Aravindan Ilangovan / Heath Murray /  Abstract: Genome duplication is essential for the proliferation of cellular life and this process is generally initiated by dedicated replication proteins at chromosome origins. In bacteria, DNA replication is ...Genome duplication is essential for the proliferation of cellular life and this process is generally initiated by dedicated replication proteins at chromosome origins. In bacteria, DNA replication is initiated by the ubiquitous DnaA protein, which assembles into an oligomeric complex at the chromosome origin (oriC) that engages both double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) to promote DNA duplex opening. However, the mechanism of DnaA specifically opening a replication origin was unknown. Here we show that Bacillus subtilis DnaA assembles into a continuous oligomer at the site of DNA melting, extending from a dsDNA anchor to engage a single DNA strand. Within this complex, two nucleobases of each ssDNA binding motif (DnaA-trio) are captured within a dinucleotide binding pocket created by adjacent DnaA proteins. These results provide a molecular basis for DnaA specifically engaging the conserved sequence elements within the bacterial chromosome origin basal unwinding system (BUS). | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16256.map.gz emd_16256.map.gz | 62.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16256-v30.xml emd-16256-v30.xml emd-16256.xml emd-16256.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16256.png emd_16256.png | 15.4 KB | ||
| Filedesc metadata |  emd-16256.cif.gz emd-16256.cif.gz | 3.9 KB | ||
| Others |  emd_16256_half_map_1.map.gz emd_16256_half_map_1.map.gz emd_16256_half_map_2.map.gz emd_16256_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16256 http://ftp.pdbj.org/pub/emdb/structures/EMD-16256 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16256 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16256 | HTTPS FTP |
-Validation report
| Summary document |  emd_16256_validation.pdf.gz emd_16256_validation.pdf.gz | 678.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16256_full_validation.pdf.gz emd_16256_full_validation.pdf.gz | 678.2 KB | Display | |
| Data in XML |  emd_16256_validation.xml.gz emd_16256_validation.xml.gz | 13.8 KB | Display | |
| Data in CIF |  emd_16256_validation.cif.gz emd_16256_validation.cif.gz | 16.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16256 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16256 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16256 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16256 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16256.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16256.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.072 Å | ||||||||||||||||||||||||||||||||||||
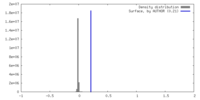
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16256_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
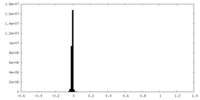
| Density Histograms |
-Half map: #2
| File | emd_16256_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
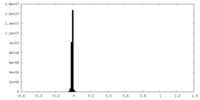
| Density Histograms |
- Sample components
Sample components
-Entire : Basal unwinding complex
| Entire | Name: Basal unwinding complex |
|---|---|
| Components |
|
-Supramolecule #1: Basal unwinding complex
| Supramolecule | Name: Basal unwinding complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / Details: Nucleoprotein complex |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: Abinitio |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 5.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 1192717 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)