[English] 日本語
 Yorodumi
Yorodumi- EMDB-16098: Cryo-EM structure of a contractile injection system in Streptomyc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a contractile injection system in Streptomyces coelicolor, the contracted sheath shell. | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | STRUCTURAL PROTEIN / Contractile injection system | ||||||||||||
| Function / homology | Phage tail sheath protein, beta-sandwich domain / Phage tail sheath protein beta-sandwich domain / : / Tail sheath protein, subtilisin-like domain / Phage tail sheath protein subtilisin-like domain / Tail sheath protein, C-terminal domain / Phage tail sheath C-terminal domain / Phage tail sheath protein Function and homology information Function and homology information | ||||||||||||
| Biological species |  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) | ||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||
 Authors Authors | Casu B / Sallmen JW / Schlimpert S / Pilhofer M | ||||||||||||
| Funding support |  Switzerland, European Union, Switzerland, European Union,  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2023 Journal: Nat Microbiol / Year: 2023Title: Cytoplasmic contractile injection systems mediate cell death in Streptomyces. Authors: Bastien Casu / Joseph W Sallmen / Susan Schlimpert / Martin Pilhofer /   Abstract: Contractile injection systems (CIS) are bacteriophage tail-like structures that mediate bacterial cell-cell interactions. While CIS are highly abundant across diverse bacterial phyla, representative ...Contractile injection systems (CIS) are bacteriophage tail-like structures that mediate bacterial cell-cell interactions. While CIS are highly abundant across diverse bacterial phyla, representative gene clusters in Gram-positive organisms remain poorly studied. Here we characterize a CIS in the Gram-positive multicellular model organism Streptomyces coelicolor and show that, in contrast to most other CIS, S. coelicolor CIS (CIS) mediate cell death in response to stress and impact cellular development. CIS are expressed in the cytoplasm of vegetative hyphae and are not released into the medium. Our cryo-electron microscopy structure enabled the engineering of non-contractile and fluorescently tagged CIS assemblies. Cryo-electron tomography showed that CIS contraction is linked to reduced cellular integrity. Fluorescence light microscopy furthermore revealed that functional CIS mediate cell death upon encountering different types of stress. The absence of functional CIS had an impact on hyphal differentiation and secondary metabolite production. Finally, we identified three putative effector proteins, which when absent, phenocopied other CIS mutants. Our results provide new functional insights into CIS in Gram-positive organisms and a framework for studying novel intracellular roles, including regulated cell death and life-cycle progression in multicellular bacteria. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16098.map.gz emd_16098.map.gz | 115.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16098-v30.xml emd-16098-v30.xml emd-16098.xml emd-16098.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16098.png emd_16098.png | 143.5 KB | ||
| Filedesc metadata |  emd-16098.cif.gz emd-16098.cif.gz | 5.3 KB | ||
| Others |  emd_16098_half_map_1.map.gz emd_16098_half_map_1.map.gz emd_16098_half_map_2.map.gz emd_16098_half_map_2.map.gz | 96.7 MB 96.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16098 http://ftp.pdbj.org/pub/emdb/structures/EMD-16098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16098 | HTTPS FTP |
-Related structure data
| Related structure data |  8bkyMC  8bl4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16098.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16098.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||
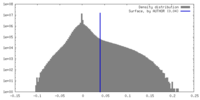
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_16098_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
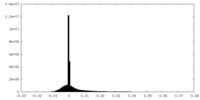
| Density Histograms |
-Half map: #1
| File | emd_16098_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The sheath structure of a contractile injection system in Strepto...
| Entire | Name: The sheath structure of a contractile injection system in Streptomyces coelicolor |
|---|---|
| Components |
|
-Supramolecule #1: The sheath structure of a contractile injection system in Strepto...
| Supramolecule | Name: The sheath structure of a contractile injection system in Streptomyces coelicolor type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
-Macromolecule #1: Phage tail sheath protein
| Macromolecule | Name: Phage tail sheath protein / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
| Molecular weight | Theoretical: 57.009609 KDa |
| Sequence | String: MPSYLSPGVY VEEVASGSRP IEGVGTSVAA FVGLAPTGPL NEPTLVTNWT QYVAAFGDFT GGYYLAHSVY GFFNNGGSAA YVVRVGGSA EDAAADGSVN GAAAPAAVTG STAKALPAAE PKQLGTFAVT ATAAGQSGPL TVEVADPEGE GPAERFKLIV K DGDKPVET ...String: MPSYLSPGVY VEEVASGSRP IEGVGTSVAA FVGLAPTGPL NEPTLVTNWT QYVAAFGDFT GGYYLAHSVY GFFNNGGSAA YVVRVGGSA EDAAADGSVN GAAAPAAVTG STAKALPAAE PKQLGTFAVT ATAAGQSGPL TVEVADPEGE GPAERFKLIV K DGDKPVET FDVSAKKGNR SYVVTQVKER SKLITVTEAA PSAQLVRPEN QSLTLPAPPS AAPAVPAGQA ESAHPGPAQY LG DSSDRTG FGGLEAIDEI SMVAVPDLMA AYQRGAIDLE AVKAVQLGLI AHCELMGDRV AIIDPPPNQN ARQIRVWRQE TAG YDSKYA ALYYPWIKSF DPATGQSRLV PPSGHVAGIW ARNDSERGVH KAPANEVVRG AVDLELQITR GEQDLLNPIG VNCI RSFPG RGIRVWGART LSSDPAWRYL NIRRYFNYLE ESILIGTQWV VFEPNDHNLW ARIRRNVSAF LVNEWRNGAL FGQSP DQAY YVKCDEETNP PESVDLGRVV CEIGIAPVKP AEFVIFRLAQ FSSGGGELDE UniProtKB: Phage tail sheath protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 38.5 Å Applied symmetry - Helical parameters - Δ&Phi: 23.10 ° Applied symmetry - Helical parameters - Axial symmetry: C6 (6 fold cyclic) Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 4854 |
|---|---|
| Startup model | Type of model: EMDB MAP EMDB ID: |
| Final angle assignment | Type: NOT APPLICABLE |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8bky: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)