[English] 日本語
 Yorodumi
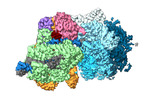
Yorodumi- EMDB-15975: Cas12k-sgRNA-dsDNA-S15-TniQ-TnsC transposon recruitment complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cas12k-sgRNA-dsDNA-S15-TniQ-TnsC transposon recruitment complex | |||||||||||||||
 Map data Map data | Cas12k transposon recruitment complex | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Cas12k / sgRNA / S15 / TniQ / TnsC / CRISPR-Cas / Tn7-like transposons / transposition / RNA BINDING PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of translation / ribosomal small subunit assembly / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytoplasmic translation / rRNA binding / structural constituent of ribosome / translation / cytoplasm / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Scytonema hofmannii (bacteria) / Scytonema hofmannii (bacteria) /  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||||||||
 Authors Authors | Schmitz M / Querques I / Oberli S / Chanez C / Jinek M | |||||||||||||||
| Funding support |  Switzerland, European Union, Switzerland, European Union,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Structural basis for the assembly of the type V CRISPR-associated transposon complex. Authors: Michael Schmitz / Irma Querques / Seraina Oberli / Christelle Chanez / Martin Jinek /  Abstract: CRISPR-Cas systems have been co-opted by Tn7-like transposable elements to direct RNA-guided transposition. Type V-K CRISPR-associated transposons rely on the concerted activities of the ...CRISPR-Cas systems have been co-opted by Tn7-like transposable elements to direct RNA-guided transposition. Type V-K CRISPR-associated transposons rely on the concerted activities of the pseudonuclease Cas12k, the AAA+ ATPase TnsC, the Zn-finger protein TniQ, and the transposase TnsB. Here we present a cryo-electron microscopic structure of a target DNA-bound Cas12k-transposon recruitment complex comprised of RNA-guided Cas12k, TniQ, a polymeric TnsC filament and, unexpectedly, the ribosomal protein S15. Complex assembly, mediated by a network of interactions involving the guide RNA, TniQ, and S15, results in R-loop completion. TniQ contacts two TnsC protomers at the Cas12k-proximal filament end, likely nucleating its polymerization. Transposition activity assays corroborate our structural findings, implying that S15 is a bona fide component of the type V crRNA-guided transposon machinery. Altogether, our work uncovers key mechanistic aspects underpinning RNA-mediated assembly of CRISPR-associated transposons to guide their development as programmable tools for site-specific insertion of large DNA payloads. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15975.map.gz emd_15975.map.gz | 338.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15975-v30.xml emd-15975-v30.xml emd-15975.xml emd-15975.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15975.png emd_15975.png | 114.7 KB | ||
| Filedesc metadata |  emd-15975.cif.gz emd-15975.cif.gz | 7.1 KB | ||
| Others |  emd_15975_half_map_1.map.gz emd_15975_half_map_1.map.gz emd_15975_half_map_2.map.gz emd_15975_half_map_2.map.gz | 339.3 MB 337.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15975 http://ftp.pdbj.org/pub/emdb/structures/EMD-15975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15975 | HTTPS FTP |
-Related structure data
| Related structure data |  8bd5MC  8bd4C  8bd6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15975.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15975.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cas12k transposon recruitment complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
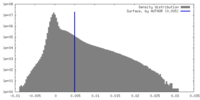
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half2
| File | emd_15975_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
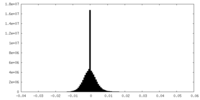
| Density Histograms |
-Half map: Half1
| File | emd_15975_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
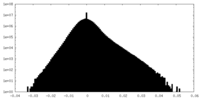
| Density Histograms |
- Sample components
Sample components
+Entire : Target DNA bound Cas12k-sgRNA-S15-TniQ-TnsC transposon recruitmen...
+Supramolecule #1: Target DNA bound Cas12k-sgRNA-S15-TniQ-TnsC transposon recruitmen...
+Macromolecule #1: ShCas12k
+Macromolecule #5: TnsC
+Macromolecule #6: TniQ (Homology model)
+Macromolecule #7: 30S ribosomal protein S15
+Macromolecule #2: sgRNA
+Macromolecule #3: DNA target strand
+Macromolecule #4: DNA non-target strand
+Macromolecule #8: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #9: MAGNESIUM ION
+Macromolecule #10: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 75 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 67.68 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1.2) / Number images used: 75000 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)