[English] 日本語
 Yorodumi
Yorodumi- EMDB-1588: Cryo-EM structure of the native GroEL-GroES complex from Thermus ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1588 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the native GroEL-GroES complex from Thermus thermophilus encapsulating substrate inside the cavity. | |||||||||
 Map data Map data | a EM-map of the GroEL-GroES-ADP-peptide complex from Thermus thermophilus | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Thermus thermophilus (bacteria) / synthetic construct (others) Thermus thermophilus (bacteria) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 23.0 Å | |||||||||
 Authors Authors | Kanno R / Koike-Takeshita A / Yokoyama K / Taguchi H / Mitsuoka K | |||||||||
 Citation Citation |  Journal: Structure / Year: 2009 Journal: Structure / Year: 2009Title: Cryo-EM structure of the native GroEL-GroES complex from thermus thermophilus encapsulating substrate inside the cavity. Authors: Ryo Kanno / Ayumi Koike-Takeshita / Ken Yokoyama / Hideki Taguchi / Kaoru Mitsuoka /  Abstract: The chaperonin GroEL interacts with various proteins, leading them to adopt their correct conformations with the aid of GroES and ATP. The actual mechanism is still being debated. In this study, by ...The chaperonin GroEL interacts with various proteins, leading them to adopt their correct conformations with the aid of GroES and ATP. The actual mechanism is still being debated. In this study, by use of cryo-electron microscopy, we determined the solution structure of the Thermus thermophilus GroEL-GroES complex encapsulating its substrate proteins. We observed the averaged density of substrate proteins in the center of the GroEL-GroES cavity. The position of the averaged substrate density in the cavity suggested a repulsive interaction between a majority of the substrate proteins and the interior wall of the cavity, which is suitable for substrate release. In addition, we observed a distortion of the cis-GroEL ring, especially at the position near the substrate, which indicated that the interaction between the encapsulated proteins and the GroEL ring results in an adjustment in the cavity's shape to accommodate the substrate. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1588.map.gz emd_1588.map.gz | 39.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1588-v30.xml emd-1588-v30.xml emd-1588.xml emd-1588.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| Images |  1588.gif 1588.gif | 107.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1588 http://ftp.pdbj.org/pub/emdb/structures/EMD-1588 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1588 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1588 | HTTPS FTP |
-Validation report
| Summary document |  emd_1588_validation.pdf.gz emd_1588_validation.pdf.gz | 233.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1588_full_validation.pdf.gz emd_1588_full_validation.pdf.gz | 232.2 KB | Display | |
| Data in XML |  emd_1588_validation.xml.gz emd_1588_validation.xml.gz | 6.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1588 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1588 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1588 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1588 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1588.map.gz / Format: CCP4 / Size: 41.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1588.map.gz / Format: CCP4 / Size: 41.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | a EM-map of the GroEL-GroES-ADP-peptide complex from Thermus thermophilus | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.63 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
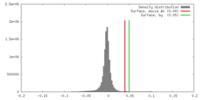
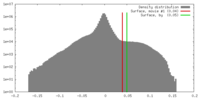
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The GroEL-GroES-ADP-peptide complex from Thermus thermophilus
| Entire | Name: The GroEL-GroES-ADP-peptide complex from Thermus thermophilus |
|---|---|
| Components |
|
-Supramolecule #1000: The GroEL-GroES-ADP-peptide complex from Thermus thermophilus
| Supramolecule | Name: The GroEL-GroES-ADP-peptide complex from Thermus thermophilus type: sample / ID: 1000 / Number unique components: 4 |
|---|---|
| Molecular weight | Theoretical: 870 KDa |
-Macromolecule #1: GroEL
| Macromolecule | Name: GroEL / type: protein_or_peptide / ID: 1 / Name.synonym: chaperonin / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
-Macromolecule #2: GroES
| Macromolecule | Name: GroES / type: protein_or_peptide / ID: 2 / Name.synonym: co-chaperonin / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
-Macromolecule #4: peptide
| Macromolecule | Name: peptide / type: protein_or_peptide / ID: 4 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #3: ADP
| Macromolecule | Name: ADP / type: ligand / ID: 3 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: METHANE / Instrument: OTHER |
|---|
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | JEOL 3200FSC |
| Image recording | Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 336 / Bits/pixel: 12 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: JEOL 3200FSC CRYOHOLDER |
- Electron microscopy #2
Electron microscopy #2
| Microscopy ID | 2 |
|---|---|
| Microscope | JEOL 2200FS |
| Image recording | Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 336 / Bits/pixel: 12 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: JEOL 3200FSC CRYOHOLDER |
- Image processing
Image processing
| CTF correction | Details: CTF correction with wiener fIlter when 2D averaging |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IMAGIC |
-Atomic model buiding 1
| Initial model | PDB ID:  1wf4 |
|---|---|
| Refinement | Space: REAL |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)