+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | p97-p37-SPI substrate complex | ||||||||||||
 Map data Map data | p97 complex with p37 adaptor and SPI substrate | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | AAA+ ATPase / p97 / VCP / Cdc48 / unfoldase / protein phosphatase-1 / protein maturation / CHAPERONE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of protein localization to centrosome / positive regulation of mitotic centrosome separation / PTW/PP1 phosphatase complex / regulation of nucleocytoplasmic transport / protein phosphatase 1 binding / nuclear membrane reassembly / lamin binding / protein phosphatase regulator activity / spindle pole centrosome / : ...negative regulation of protein localization to centrosome / positive regulation of mitotic centrosome separation / PTW/PP1 phosphatase complex / regulation of nucleocytoplasmic transport / protein phosphatase 1 binding / nuclear membrane reassembly / lamin binding / protein phosphatase regulator activity / spindle pole centrosome / : / flavin adenine dinucleotide catabolic process / VCP-NSFL1C complex / endosome to lysosome transport via multivesicular body sorting pathway / endoplasmic reticulum stress-induced pre-emptive quality control / BAT3 complex binding / SHOC2 M1731 mutant abolishes MRAS complex function / Gain-of-function MRAS complexes activate RAF signaling / cellular response to arsenite ion / protein-DNA covalent cross-linking repair / Derlin-1 retrotranslocation complex / positive regulation of protein K63-linked deubiquitination / cytoplasm protein quality control / positive regulation of oxidative phosphorylation / : / aggresome assembly / deubiquitinase activator activity / mitotic spindle disassembly / ubiquitin-modified protein reader activity / regulation of protein localization to chromatin / VCP-NPL4-UFD1 AAA ATPase complex / cellular response to misfolded protein / negative regulation of protein localization to chromatin / positive regulation of mitochondrial membrane potential / vesicle-fusing ATPase / K48-linked polyubiquitin modification-dependent protein binding / regulation of aerobic respiration / retrograde protein transport, ER to cytosol / stress granule disassembly / glycogen metabolic process / protein-serine/threonine phosphatase / ATPase complex / Triglyceride catabolism / entrainment of circadian clock by photoperiod / regulation of synapse organization / ubiquitin-specific protease binding / Golgi organization / Maturation of hRSV A proteins / protein serine/threonine phosphatase activity / phosphatase activity / microtubule organizing center / positive regulation of ATP biosynthetic process / MHC class I protein binding / cleavage furrow / ubiquitin-like protein ligase binding / establishment of mitotic spindle orientation / phosphoprotein phosphatase activity / mitotic sister chromatid segregation / RHOH GTPase cycle / blastocyst development / positive regulation of glial cell proliferation / polyubiquitin modification-dependent protein binding / autophagosome assembly / autophagosome maturation / negative regulation of hippo signaling / endoplasmic reticulum to Golgi vesicle-mediated transport / HSF1 activation / translesion synthesis / interstrand cross-link repair / protein dephosphorylation / ATP metabolic process / proteasomal protein catabolic process / endoplasmic reticulum unfolded protein response / Protein methylation / enzyme regulator activity / Attachment and Entry / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / ERAD pathway / lipid droplet / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / proteasome complex / Resolution of Sister Chromatid Cohesion / viral genome replication / Downregulation of TGF-beta receptor signaling / Josephin domain DUBs / ubiquitin binding / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / negative regulation of smoothened signaling pathway / macroautophagy / positive regulation of protein-containing complex assembly / circadian regulation of gene expression / Hh mutants are degraded by ERAD / establishment of protein localization / Hedgehog ligand biogenesis / RHO GTPases Activate Formins / Defective CFTR causes cystic fibrosis / RAF activation / positive regulation of non-canonical NF-kappaB signal transduction / Translesion Synthesis by POLH / regulation of circadian rhythm Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
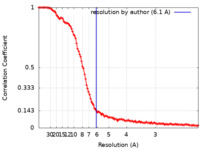
| Method | single particle reconstruction / cryo EM / Resolution: 6.1 Å | ||||||||||||
 Authors Authors | van den Boom J / Marini G / Meyer H / Saibil H | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: EMBO J / Year: 2023 Journal: EMBO J / Year: 2023Title: Structural basis of ubiquitin-independent PP1 complex disassembly by p97. Authors: Johannes van den Boom / Guendalina Marini / Hemmo Meyer / Helen R Saibil /   Abstract: The AAA+-ATPase p97 (also called VCP or Cdc48) unfolds proteins and disassembles protein complexes in numerous cellular processes, but how substrate complexes are loaded onto p97 and disassembled is ...The AAA+-ATPase p97 (also called VCP or Cdc48) unfolds proteins and disassembles protein complexes in numerous cellular processes, but how substrate complexes are loaded onto p97 and disassembled is unclear. Here, we present cryo-EM structures of p97 in the process of disassembling a protein phosphatase-1 (PP1) complex by extracting an inhibitory subunit from PP1. We show that PP1 and its partners SDS22 and inhibitor-3 (I3) are loaded tightly onto p97, surprisingly via a direct contact of SDS22 with the p97 N-domain. Loading is assisted by the p37 adapter that bridges two adjacent p97 N-domains underneath the substrate complex. A stretch of I3 is threaded into the central channel of the spiral-shaped p97 hexamer, while other elements of I3 are still attached to PP1. Thus, our data show how p97 arranges a protein complex between the p97 N-domain and central channel, suggesting a hold-and-extract mechanism for p97-mediated disassembly. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15861.map.gz emd_15861.map.gz | 116.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15861-v30.xml emd-15861-v30.xml emd-15861.xml emd-15861.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15861_fsc.xml emd_15861_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_15861.png emd_15861.png | 166.6 KB | ||
| Filedesc metadata |  emd-15861.cif.gz emd-15861.cif.gz | 7 KB | ||
| Others |  emd_15861_half_map_1.map.gz emd_15861_half_map_1.map.gz emd_15861_half_map_2.map.gz emd_15861_half_map_2.map.gz | 98.7 MB 98.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15861 http://ftp.pdbj.org/pub/emdb/structures/EMD-15861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15861 | HTTPS FTP |
-Validation report
| Summary document |  emd_15861_validation.pdf.gz emd_15861_validation.pdf.gz | 901.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15861_full_validation.pdf.gz emd_15861_full_validation.pdf.gz | 901.2 KB | Display | |
| Data in XML |  emd_15861_validation.xml.gz emd_15861_validation.xml.gz | 18.4 KB | Display | |
| Data in CIF |  emd_15861_validation.cif.gz emd_15861_validation.cif.gz | 24.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15861 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15861 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15861 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15861 | HTTPS FTP |
-Related structure data
| Related structure data |  8b5rMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15861.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15861.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | p97 complex with p37 adaptor and SPI substrate | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
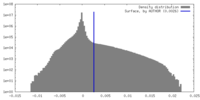
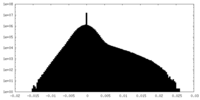
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 1
| File | emd_15861_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
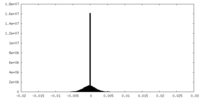
| Density Histograms |
-Half map: Half map 2
| File | emd_15861_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
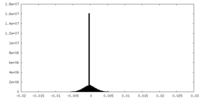
| Density Histograms |
- Sample components
Sample components
-Entire : p97 complex with p37 adaptor and SPI substrate
| Entire | Name: p97 complex with p37 adaptor and SPI substrate |
|---|---|
| Components |
|
-Supramolecule #1: p97 complex with p37 adaptor and SPI substrate
| Supramolecule | Name: p97 complex with p37 adaptor and SPI substrate / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4, #6, #5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transitional endoplasmic reticulum ATPase
| Macromolecule | Name: Transitional endoplasmic reticulum ATPase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: vesicle-fusing ATPase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 90.265695 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHASG ADSKGDDLST AILKQKNRPN RLIVDEAINE DNSVVSLSQP KMDELQLFRG DTVLLKGKKR REAVCIVLSD DTCSDEKIR MNRVVRNNLR VRLGDVISIQ PCPDVKYGKR IHVLPIDDTV EGITGNLFEV YLKPYFLEAY RPIRKGDIFL V RGGMRAVE ...String: MHHHHHHASG ADSKGDDLST AILKQKNRPN RLIVDEAINE DNSVVSLSQP KMDELQLFRG DTVLLKGKKR REAVCIVLSD DTCSDEKIR MNRVVRNNLR VRLGDVISIQ PCPDVKYGKR IHVLPIDDTV EGITGNLFEV YLKPYFLEAY RPIRKGDIFL V RGGMRAVE FKVVETDPSP YCIVAPDTVI HCEGEPIKRE DEEESLNEVG YDDIGGCRKQ LAQIKEMVEL PLRHPALFKA IG VKPPRGI LLYGPPGTGK TLIARAVANE TGAFFFLING PEIMSKLAGE SESNLRKAFE EAEKNAPAII FIDELDAIAP KRE KTHGEV ERRIVSQLLT LMDGLKQRAH VIVMAATNRP NSIDPALRRF GRFDREVDIG IPDATGRLEI LQIHTKNMKL ADDV DLEQV ANETHGHVGA DLAALCSEAA LQAIRKKMDL IDLEDETIDA EVMNSLAVTM DDFRWALSQS NPSALRETVV EVPQV TWED IGGLEDVKRE LQELVQYPVE HPDKFLKFGM TPSKGVLFYG PPGCGKTLLA KAIANECQAN FISIKGPELL TMWFGE SEA NVREIFDKAR QAAPCVLFFD ELDSIAKARG GNIGDGGGAA DRVINQILTE MDGMSTKKNV FIIGATNRPD IIDPAIL RP GRLDQLIYIP LPDEKSRVAI LKANLRKSPV AKDVDLEFLA KMTNGFSGAD LTEICQRACK LAIRESIESE IRRERERQ T NPSAMEVEED DPVPEIRRDH FEEAMRFARR SVSDNDIRKY EMFAQTLQQS RGFGSFRFPS GNQGGAGPSQ GSGGGTGGS VYTEDNDDDL YG UniProtKB: Transitional endoplasmic reticulum ATPase |
-Macromolecule #2: I3 sequence being threaded through the p97 channel
| Macromolecule | Name: I3 sequence being threaded through the p97 channel / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.890321 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK) |
-Macromolecule #3: Serine/threonine-protein phosphatase PP1-gamma catalytic subunit
| Macromolecule | Name: Serine/threonine-protein phosphatase PP1-gamma catalytic subunit type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: protein-serine/threonine phosphatase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.030777 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MADLDKLNID SIIQRLLEVR GSKPGKNVQL QENEIRGLCL KSREIFLSQP ILLELEAPLK ICGDIHGQYY DLLRLFEYGG FPPESNYLF LGDYVDRGKQ SLETICLLLA YKIKYPENFF LLRGNHECAS INRIYGFYDE CKRRYNIKLW KTFTDCFNCL P IAAIVDEK ...String: MADLDKLNID SIIQRLLEVR GSKPGKNVQL QENEIRGLCL KSREIFLSQP ILLELEAPLK ICGDIHGQYY DLLRLFEYGG FPPESNYLF LGDYVDRGKQ SLETICLLLA YKIKYPENFF LLRGNHECAS INRIYGFYDE CKRRYNIKLW KTFTDCFNCL P IAAIVDEK IFCCHGGLSP DLQSMEQIRR IMRPTDVPDQ GLLCDLLWSD PDKDVLGWGE NDRGVSFTFG AEVVAKFLHK HD LDLICRA HQVVEDGYEF FAKRQLVTLF SAPNYCGEFD NAGAMMSVDE TLMCSFQILK PAEKKKPNAT RPVTPPRGMI TKQ AKK UniProtKB: Serine/threonine-protein phosphatase PP1-gamma catalytic subunit |
-Macromolecule #4: Protein phosphatase 1 regulatory subunit 7
| Macromolecule | Name: Protein phosphatase 1 regulatory subunit 7 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.616129 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAAERGAGQQ QSQEMMEVDR RVESEESGDE EGKKHSSGIV ADLSEQSLKD GEERGEEDPE EEHELPVDME TINLDRDAED VDLNHYRIG KIEGFEVLKK VKTLCLRQNL IKCIENLEEL QSLRELDLYD NQIKKIENLE ALTELEILDI SFNLLRNIEG V DKLTRLKK ...String: MAAERGAGQQ QSQEMMEVDR RVESEESGDE EGKKHSSGIV ADLSEQSLKD GEERGEEDPE EEHELPVDME TINLDRDAED VDLNHYRIG KIEGFEVLKK VKTLCLRQNL IKCIENLEEL QSLRELDLYD NQIKKIENLE ALTELEILDI SFNLLRNIEG V DKLTRLKK LFLVNNKISK IENLSNLHQL QMLELGSNRI RAIENIDTLT NLESLFLGKN KITKLQNLDA LTNLTVLSMQ SN RLTKIEG LQNLVNLREL YLSHNGIEVI EGLENNNKLT MLDIASNRIK KIENISHLTE LQEFWMNDNL LESWSDLDEL KGA RSLETV YLERNPLQKD PQYRRKVMLA LPSVRQIDAT FVRF UniProtKB: Protein phosphatase 1 regulatory subunit 7 |
-Macromolecule #5: UBX domain-containing protein 2B
| Macromolecule | Name: UBX domain-containing protein 2B / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.123238 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FSGEGQKLGS L UniProtKB: UBX domain-containing protein 2B |
-Macromolecule #6: UBX domain-containing protein 2B
| Macromolecule | Name: UBX domain-containing protein 2B / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.75922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AVVLIDDSVP TTKIQIRLAD GSRLIQRFNS THRILDVRNF IVQSRPEFAA LDFILVTSFP NKELTDESLT LLEADILNTV LLQQLK UniProtKB: UBX domain-containing protein 2B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 3.0 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.3000000000000003 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8b5r: |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)