+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | IstA transposase cleaved donor complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA Transposition / Transposase / Cleaved donor complex / DDE domain / IS21 / IstA / Insertion sequence / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtransposition / DNA strand exchange activity / DNA integration / DNA binding Similarity search - Function | |||||||||
| Biological species |   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) | |||||||||
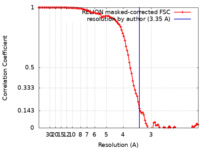
| Method | single particle reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||
 Authors Authors | Spinola-Amilibia M / de la Gandara A / Araujo-Bazan L / Berger JM / Arias-Palomo E | |||||||||
| Funding support |  Spain, 2 items Spain, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: IS21 family transposase cleaved donor complex traps two right-handed superhelical crossings. Authors: Mercedes Spínola-Amilibia / Lidia Araújo-Bazán / Álvaro de la Gándara / James M Berger / Ernesto Arias-Palomo /   Abstract: Transposases are ubiquitous enzymes that catalyze DNA rearrangement events with broad impacts on gene expression, genome evolution, and the spread of drug-resistance in bacteria. Here, we use ...Transposases are ubiquitous enzymes that catalyze DNA rearrangement events with broad impacts on gene expression, genome evolution, and the spread of drug-resistance in bacteria. Here, we use biochemical and structural approaches to define the molecular determinants by which IstA, a transposase present in the widespread IS21 family of mobile elements, catalyzes efficient DNA transposition. Solution studies show that IstA engages the transposon terminal sequences to form a high-molecular weight complex and promote DNA integration. A 3.4 Å resolution structure of the transposase bound to transposon ends corroborates our biochemical findings and reveals that IstA self-assembles into a highly intertwined tetramer that synapses two supercoiled terminal inverted repeats. The three-dimensional organization of the IstA•DNA cleaved donor complex reveals remarkable similarities with retroviral integrases and classic transposase systems, such as Tn7 and bacteriophage Mu, and provides insights into IS21 transposition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15848.map.gz emd_15848.map.gz | 85.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15848-v30.xml emd-15848-v30.xml emd-15848.xml emd-15848.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15848_fsc.xml emd_15848_fsc.xml | 10.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_15848.png emd_15848.png | 72.8 KB | ||
| Masks |  emd_15848_msk_1.map emd_15848_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15848.cif.gz emd-15848.cif.gz | 6.9 KB | ||
| Others |  emd_15848_additional_1.map.gz emd_15848_additional_1.map.gz emd_15848_half_map_1.map.gz emd_15848_half_map_1.map.gz emd_15848_half_map_2.map.gz emd_15848_half_map_2.map.gz | 69.9 MB 70.1 MB 70.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15848 http://ftp.pdbj.org/pub/emdb/structures/EMD-15848 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15848 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15848 | HTTPS FTP |
-Related structure data
| Related structure data |  8b4hMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15848.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15848.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15848_msk_1.map emd_15848_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_15848_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15848_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15848_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : IstA transposase cleaved donor complex
| Entire | Name: IstA transposase cleaved donor complex |
|---|---|
| Components |
|
-Supramolecule #1: IstA transposase cleaved donor complex
| Supramolecule | Name: IstA transposase cleaved donor complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Complex of IstA transposase bound to two right IS5376 transposon ends |
|---|---|
| Source (natural) | Organism:   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) |
| Molecular weight | Theoretical: 225 KDa |
-Macromolecule #1: Putative transposase for insertion sequence element IS5376
| Macromolecule | Name: Putative transposase for insertion sequence element IS5376 type: protein_or_peptide / ID: 1 Details: The residues annotated as cloning artefact are the following five flanking bases of the X67861 gene Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) |
| Molecular weight | Theoretical: 47.687723 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MITRGEFFMI KEMYERGMSI SDIARELGID RKTVRKYIHS PNPPSKSKRK QRKSKLDPFK PYLQKRMLED GVFNSEKLFF EIRQQGYTG GKTILKDYMK PFRETAKKKY TVRYETLPGE QMQVDWKEVG EVVIEGKKVK LSLFVATLGY SRMKYAVFTT S QDQEHLME ...String: MITRGEFFMI KEMYERGMSI SDIARELGID RKTVRKYIHS PNPPSKSKRK QRKSKLDPFK PYLQKRMLED GVFNSEKLFF EIRQQGYTG GKTILKDYMK PFRETAKKKY TVRYETLPGE QMQVDWKEVG EVVIEGKKVK LSLFVATLGY SRMKYAVFTT S QDQEHLME CLIQSFKYFG GVPKKVLFDN MKTVTDGREQ GVVKWNQRFS EFASYYGFIP KVCRPYRAQT KGKVERAIQY IM DHFYVGT AFESIEELNF LLHRWLDQVA NRKPNATTGI SPQERWAEES LKPLPLKDYD TSYLSYRKVH WDGSFSYKGE QWL LSAEYA GKEILVKERL NGDIRLYFRG EEISHVDQQK KVISFAEKIK KKQTEMAATI SPVSVEVDTR PLSVYDAFLR GESS ENLYF Q UniProtKB: Putative transposase for insertion sequence element IS5376 |
-Macromolecule #2: DNA (57-MER) / right IS21 transposon end (insertion sequence IS5376)
| Macromolecule | Name: DNA (57-MER) / right IS21 transposon end (insertion sequence IS5376) type: dna / ID: 2 / Details: GB X67861 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) |
| Molecular weight | Theoretical: 18.384814 KDa |
| Sequence | String: (DA)(DT)(DT)(DC)(DA)(DT)(DG)(DT)(DC)(DA) (DA)(DG)(DG)(DC)(DC)(DG)(DA)(DT)(DT)(DA) (DT)(DT)(DT)(DT)(DT)(DT)(DC)(DC)(DC) (DC)(DA)(DA)(DA)(DA)(DT)(DC)(DG)(DC)(DC) (DG) (DG)(DT)(DT)(DT)(DA)(DA) ...String: (DA)(DT)(DT)(DC)(DA)(DT)(DG)(DT)(DC)(DA) (DA)(DG)(DG)(DC)(DC)(DG)(DA)(DT)(DT)(DA) (DT)(DT)(DT)(DT)(DT)(DT)(DC)(DC)(DC) (DC)(DA)(DA)(DA)(DA)(DT)(DC)(DG)(DC)(DC) (DG) (DG)(DT)(DT)(DT)(DA)(DA)(DA)(DA) (DT)(DT)(DC)(DC)(DC)(DC)(DA)(DG)(DA)(DA) (DG)(DG) |
-Macromolecule #3: DNA (55-MER) / right IS21 transposon end (insertion sequence IS5376)
| Macromolecule | Name: DNA (55-MER) / right IS21 transposon end (insertion sequence IS5376) type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) |
| Molecular weight | Theoretical: 17.029939 KDa |
| Sequence | String: (DC)(DC)(DT)(DT)(DC)(DT)(DG)(DG)(DG)(DG) (DA)(DA)(DT)(DT)(DT)(DT)(DA)(DA)(DA)(DC) (DC)(DG)(DG)(DC)(DG)(DA)(DT)(DT)(DT) (DT)(DG)(DG)(DG)(DG)(DA)(DA)(DA)(DA)(DA) (DA) (DT)(DA)(DA)(DT)(DC)(DG) ...String: (DC)(DC)(DT)(DT)(DC)(DT)(DG)(DG)(DG)(DG) (DA)(DA)(DT)(DT)(DT)(DT)(DA)(DA)(DA)(DC) (DC)(DG)(DG)(DC)(DG)(DA)(DT)(DT)(DT) (DT)(DG)(DG)(DG)(DG)(DA)(DA)(DA)(DA)(DA) (DA) (DT)(DA)(DA)(DT)(DC)(DG)(DG)(DC) (DC)(DT)(DT)(DG)(DA)(DC)(DA) GENBANK: GENBANK: X67861.1 |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.119 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: 25 mA | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 3.36 sec. / Average electron dose: 59.7 e/Å2 / Details: Single shot per hole |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 115.818 |
|---|---|
| Output model |  PDB-8b4h: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)