+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Chaetoceros socialis forma radians RNA virus 1 full capsid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pseudo-T=3 / icosahedral / VIRUS | |||||||||
| Function / homology | Capsid protein VP4, dicistrovirus / Cricket paralysis virus, VP4 / Dicistrovirus, capsid-polyprotein, C-terminal / CRPV capsid protein like / virion component / Picornavirus/Calicivirus coat protein / Viral coat protein subunit / Structural polyprotein Function and homology information Function and homology information | |||||||||
| Biological species |  Chaetoceros socialis forma radians RNA virus 1 Chaetoceros socialis forma radians RNA virus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Wang H / Okamoto K / Munke A | |||||||||
| Funding support |  Sweden, 1 items Sweden, 1 items
| |||||||||
 Citation Citation |  Journal: Viruses / Year: 2022 Journal: Viruses / Year: 2022Title: Structural Insights into Common and Host-Specific Receptor-Binding Mechanisms in Algal Picorna-like Viruses. Authors: Han Wang / Anna Munke / Siqi Li / Yuji Tomaru / Kenta Okamoto /    Abstract: viruses are abundant algal viruses that regulate the dynamics of algal blooms in aquatic environments. They employ a narrow host range because they merely lyse their algal host species. This host- ... viruses are abundant algal viruses that regulate the dynamics of algal blooms in aquatic environments. They employ a narrow host range because they merely lyse their algal host species. This host-specific lysis is thought to correspond to the unique receptor-binding mechanism of the viruses. Here, we present the atomic structures of the full and empty capsids of Chaetoceros socialis forma radians RNA virus 1 built-in 3.0 Å and 3.1 Å cryo-electron microscopy maps. The empty capsid structure and the structural variability provide insights into its assembly and uncoating intermediates. In conjunction with the previously reported atomic model of the Chaetoceros tenuissimus RNA virus type II capsid, we have identified the common and diverse structural features of the VP1 surface between the viruses. We have also tested the potential usage of AlphaFold2 for structural prediction of the VP1s and a subsequent structural phylogeny for classifying viruses by their hosts. These findings will be crucial for inferring the host-specific receptor-binding mechanism in viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15823.map.gz emd_15823.map.gz | 154.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15823-v30.xml emd-15823-v30.xml emd-15823.xml emd-15823.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15823.png emd_15823.png | 224.3 KB | ||
| Filedesc metadata |  emd-15823.cif.gz emd-15823.cif.gz | 5.4 KB | ||
| Others |  emd_15823_half_map_1.map.gz emd_15823_half_map_1.map.gz emd_15823_half_map_2.map.gz emd_15823_half_map_2.map.gz | 151.3 MB 151.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15823 http://ftp.pdbj.org/pub/emdb/structures/EMD-15823 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15823 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15823 | HTTPS FTP |
-Validation report
| Summary document |  emd_15823_validation.pdf.gz emd_15823_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15823_full_validation.pdf.gz emd_15823_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_15823_validation.xml.gz emd_15823_validation.xml.gz | 14.8 KB | Display | |
| Data in CIF |  emd_15823_validation.cif.gz emd_15823_validation.cif.gz | 17.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15823 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15823 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15823 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15823 | HTTPS FTP |
-Related structure data
| Related structure data |  8b38MC  8b3jC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15823.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15823.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
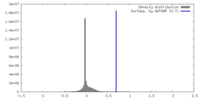
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_15823_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
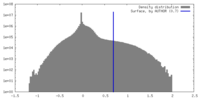
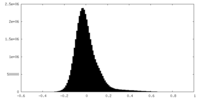
| Density Histograms |
-Half map: #2
| File | emd_15823_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
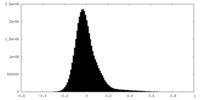
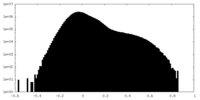
| Density Histograms |
- Sample components
Sample components
-Entire : Chaetoceros socialis forma radians RNA virus 1
| Entire | Name:  Chaetoceros socialis forma radians RNA virus 1 Chaetoceros socialis forma radians RNA virus 1 |
|---|---|
| Components |
|
-Supramolecule #1: Chaetoceros socialis forma radians RNA virus 1
| Supramolecule | Name: Chaetoceros socialis forma radians RNA virus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2169725 Sci species name: Chaetoceros socialis forma radians RNA virus 1 Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Structural polyprotein
| Macromolecule | Name: Structural polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetoceros socialis forma radians RNA virus 1 Chaetoceros socialis forma radians RNA virus 1 |
| Molecular weight | Theoretical: 29.930619 KDa |
| Sequence | String: SPLAIAGNRA GGSGAITLEP FGSDNTSPEL SKLHFGETYD HMRVLVKGYN FYKAVLDNTT ITDPDNPSGT VVIRSTVPDF PAPRGHIPN GPDKPFLPHP ENAALCTHLT WFSPCYVARR GGIRWKYLHF GARFGATDAP KGLGFVNRVP QVPYGRRPLG N ERLPLTDV ...String: SPLAIAGNRA GGSGAITLEP FGSDNTSPEL SKLHFGETYD HMRVLVKGYN FYKAVLDNTT ITDPDNPSGT VVIRSTVPDF PAPRGHIPN GPDKPFLPHP ENAALCTHLT WFSPCYVARR GGIRWKYLHF GARFGATDAP KGLGFVNRVP QVPYGRRPLG N ERLPLTDV NGFNPTELSQ AFSNENGGVY ATDLDVQPAI EVELPFYSSL RFVNPRWDDY EKIGIHRHSI DLLSYRTNGA KC EDAWMAY VAAGDDFNLS WMLSCPPFRL VNL UniProtKB: Structural polyprotein |
-Macromolecule #2: Structural polyprotein
| Macromolecule | Name: Structural polyprotein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetoceros socialis forma radians RNA virus 1 Chaetoceros socialis forma radians RNA virus 1 |
| Molecular weight | Theoretical: 26.871293 KDa |
| Sequence | String: AVKDTIEGNS ETLSGTHQNE TLALYSNVDQ TAVKIMSSID PTRADCVSND HELGNFLSRP VRIMRESISL DERTSTTIAP WDVYLRHPM INKKIANYEY LRANLVLEVV VNGGPFFYGK MLLGYTPFGY EDSLKNFNRI PIGHQNTMLS QQPHVKIDFC E STGGVLHL ...String: AVKDTIEGNS ETLSGTHQNE TLALYSNVDQ TAVKIMSSID PTRADCVSND HELGNFLSRP VRIMRESISL DERTSTTIAP WDVYLRHPM INKKIANYEY LRANLVLEVV VNGGPFFYGK MLLGYTPFGY EDSLKNFNRI PIGHQNTMLS QQPHVKIDFC E STGGVLHL PFVYNRNYMR ISEGSGEPAS MGELRLNTLN ALKNISFTGP ASSVATITVF AYLDNVELVA PSANDPITAQ QP EL UniProtKB: Structural polyprotein |
-Macromolecule #3: Structural polyprotein
| Macromolecule | Name: Structural polyprotein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetoceros socialis forma radians RNA virus 1 Chaetoceros socialis forma radians RNA virus 1 |
| Molecular weight | Theoretical: 31.098668 KDa |
| Sequence | String: CRPVVIDPPH KYRPTYVGNM ANADIAEAVD KLSLTSKQEL TINHDVIGKK SDGDDMHLST FFGREAYMDR FEWKTTDSYD TLLFYTHVH PILFKRFEAT SGDYDVGMLL PPVGYATIPF SFWRGGMTFR FSIVASAFHR GRLRIVYQPQ GGLGTVPGFS A AFNRVIDL ...String: CRPVVIDPPH KYRPTYVGNM ANADIAEAVD KLSLTSKQEL TINHDVIGKK SDGDDMHLST FFGREAYMDR FEWKTTDSYD TLLFYTHVH PILFKRFEAT SGDYDVGMLL PPVGYATIPF SFWRGGMTFR FSIVASAFHR GRLRIVYQPQ GGLGTVPGFS A AFNRVIDL GDARDFEVTV EWNQNIAFRE VHTTGSNVPS AQYTPGLDVG RTSQLPLGDQ TSVSNGVLAV YVVNDLVSPD GG TDESVEV NWFVKGAPSF EVASRDTKFA RWSTHWSQEE F UniProtKB: Structural polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 3984 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)