[English] 日本語
 Yorodumi
Yorodumi- EMDB-15777: Cryo-EM structure of ALC1 bound to an asymmetric, site-specifical... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
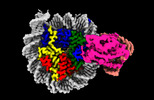
| Title | Cryo-EM structure of ALC1 bound to an asymmetric, site-specifically PARylated nucleosome | ||||||||||||
 Map data Map data | Map from non-uniform refinement of 212 256 particles in cryoSPARC. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ALC1 / CHD1L / nucleosome / PARylation / ADP-ribosylation / post-translational modification / chromatin / DNA BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationpoly-ADP-D-ribose modification-dependent protein binding / ATP-dependent chromatin remodeler activity / histone reader activity / site of DNA damage / nucleosome binding / DNA helicase activity / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / Dual Incision in GG-NER / Formation of Incision Complex in GG-NER / structural constituent of chromatin ...poly-ADP-D-ribose modification-dependent protein binding / ATP-dependent chromatin remodeler activity / histone reader activity / site of DNA damage / nucleosome binding / DNA helicase activity / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / Dual Incision in GG-NER / Formation of Incision Complex in GG-NER / structural constituent of chromatin / heterochromatin formation / nucleosome / nucleosome assembly / site of double-strand break / chromatin remodeling / protein heterodimerization activity / nucleotide binding / DNA repair / DNA damage response / ATP hydrolysis activity / DNA binding / nucleoplasm / ATP binding / nucleus Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) / | ||||||||||||
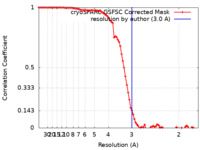
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Bacic L / Gaullier G / Deindl S | ||||||||||||
| Funding support | European Union,  Sweden, 3 items Sweden, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Asymmetric nucleosome PARylation at DNA breaks mediates directional nucleosome sliding by ALC1. Authors: Luka Bacic / Guillaume Gaullier / Jugal Mohapatra / Guanzhong Mao / Klaus Brackmann / Mikhail Panfilov / Glen Liszczak / Anton Sabantsev / Sebastian Deindl /   Abstract: The chromatin remodeler ALC1 is activated by DNA damage-induced poly(ADP-ribose) deposited by PARP1/PARP2 and their co-factor HPF1. ALC1 has emerged as a cancer drug target, but how it is recruited ...The chromatin remodeler ALC1 is activated by DNA damage-induced poly(ADP-ribose) deposited by PARP1/PARP2 and their co-factor HPF1. ALC1 has emerged as a cancer drug target, but how it is recruited to ADP-ribosylated nucleosomes to affect their positioning near DNA breaks is unknown. Here we find that PARP1/HPF1 preferentially initiates ADP-ribosylation on the histone H2B tail closest to the DNA break. To dissect the consequences of such asymmetry, we generate nucleosomes with a defined ADP-ribosylated H2B tail on one side only. The cryo-electron microscopy structure of ALC1 bound to such an asymmetric nucleosome indicates preferential engagement on one side. Using single-molecule FRET, we demonstrate that this asymmetric recruitment gives rise to directed sliding away from the DNA linker closest to the ADP-ribosylation site. Our data suggest a mechanism by which ALC1 slides nucleosomes away from a DNA break to render it more accessible to repair factors. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15777.map.gz emd_15777.map.gz | 32 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15777-v30.xml emd-15777-v30.xml emd-15777.xml emd-15777.xml | 28 KB 28 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15777_fsc.xml emd_15777_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_15777.png emd_15777.png | 81.7 KB | ||
| Masks |  emd_15777_msk_1.map emd_15777_msk_1.map emd_15777_msk_2.map emd_15777_msk_2.map | 64 MB 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15777.cif.gz emd-15777.cif.gz | 7.7 KB | ||
| Others |  emd_15777_additional_1.map.gz emd_15777_additional_1.map.gz emd_15777_half_map_1.map.gz emd_15777_half_map_1.map.gz emd_15777_half_map_2.map.gz emd_15777_half_map_2.map.gz | 54.5 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15777 http://ftp.pdbj.org/pub/emdb/structures/EMD-15777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15777 | HTTPS FTP |
-Related structure data
| Related structure data |  8b0aMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15777.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15777.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map from non-uniform refinement of 212 256 particles in cryoSPARC. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8617 Å | ||||||||||||||||||||||||||||||||||||
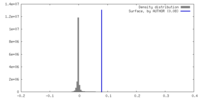
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15777_msk_1.map emd_15777_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
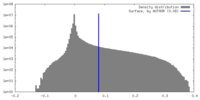
| Density Histograms |
-Mask #2
| File |  emd_15777_msk_2.map emd_15777_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
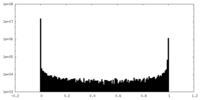
| Density Histograms |
-Additional map: Main map denoised with deepEMhancer, using the "highRes"...
| File | emd_15777_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map denoised with deepEMhancer, using the "highRes" denoising model. | ||||||||||||
| Projections & Slices |
| ||||||||||||
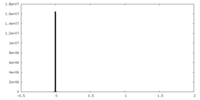
| Density Histograms |
-Half map: Half-map A from non-uniform refinement in cryoSPARC.
| File | emd_15777_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A from non-uniform refinement in cryoSPARC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map B from non-uniform refinement in cryoSPARC.
| File | emd_15777_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B from non-uniform refinement in cryoSPARC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between ALC1 and a site-specifically PARylated nucleosome
| Entire | Name: Complex between ALC1 and a site-specifically PARylated nucleosome |
|---|---|
| Components |
|
-Supramolecule #1: Complex between ALC1 and a site-specifically PARylated nucleosome
| Supramolecule | Name: Complex between ALC1 and a site-specifically PARylated nucleosome type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Site-specific ADP-ribosylation was introduced as described in https://doi.org/10.7554/elife.71502 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 304.57 KDa |
-Macromolecule #1: Chromodomain-helicase-DNA-binding protein 1-like
| Macromolecule | Name: Chromodomain-helicase-DNA-binding protein 1-like / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 98.906766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFLLRLHTEG RAEAARVQEQ DLRQWGLTGI HLRSYQLEGV NWLAQRFHCQ NGCILGDEMG LGKTCQTIAL FIYLAGRLND EGPFLILCP LSVLSNWKEE MQRFAPGLSC VTYAGDKEER ACLQQDLKQE SRFHVLLTTY EICLKDASFL KSFPWSVLVV D EAHRLKNQ ...String: MFLLRLHTEG RAEAARVQEQ DLRQWGLTGI HLRSYQLEGV NWLAQRFHCQ NGCILGDEMG LGKTCQTIAL FIYLAGRLND EGPFLILCP LSVLSNWKEE MQRFAPGLSC VTYAGDKEER ACLQQDLKQE SRFHVLLTTY EICLKDASFL KSFPWSVLVV D EAHRLKNQ SSLLHKTLSE FSVVFSLLLT GTPIQNSLQE LYSLLSFVEP DLFSKEEVGD FIQRYQDIEK ESESASELHK LL QPFLLRR VKAEVATELP KKTEVVIYHG MSALQKKYYK AILMKDLDAF ENETAKKVKL QNILSQLRKC VDHPYLFDGV EPE PFEVGD HLTEASGKLH LLDKLLAFLY SGGHRVLLFS QMTQMLDILQ DYMDYRGYSY ERVDGSVRGE ERHLAIKNFG QQPI FVFLL STRAGGVGMN LTAADTVIFV DSDFNPQNDL QAAARAHRIG QNKSVKVIRL IGRDTVEEIV YRKAASKLQL TNMII EGGH FTLGAQKPAA DADLQLSEIL KFGLDKLLAS EGSTMDEIDL ESILGETKDG QWVSDALPAA EGGSRDQEEG KNHMYL FEG KDYSKEPSKE DRKSFEQLVN LQKTLLEKAS QEGRSLRNKG SVLIPGLVEG STKRKRVLSP EELEDRQKKR QEAAAKR RR LIEEKKRQKE EAEHKKKMAW WESNNYQSFC LPSEESEPED LENGEESSAE LDYQDPDATS LKYVSGDVTH PQAGAEDA L IVHCVDDSGH WGRGGLFTAL EKRSAEPRKI YELAGKMKDL SLGGVLLFPV DDKESRNKGQ DLLALIVAQH RDRSNVLSG IKMAALEEGL KKIFLAAKKK KASVHLPRIG HATKGFNWYG TERLIRKHLA ARGIPTYIYY FPRSKAHHHH HH UniProtKB: Chromodomain-helicase-DNA-binding protein 1-like |
-Macromolecule #2: Histone H3
| Macromolecule | Name: Histone H3 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 15.403062 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARTKQTARK STGGKAPRKQ LATKAARKSA PATGGVKKPH RYRPGTVALR EIRRYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSSA VMALQEASEA YLVALFEDTN LAAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3 |
-Macromolecule #3: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 11.394426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRDA VTYTEHAKRK TVTAMDVVY ALKRQGRTLY GFGG UniProtKB: Histone H4 |
-Macromolecule #4: Histone H2A type 1
| Macromolecule | Name: Histone H2A type 1 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 14.109436 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKQGGK TRAKAKTRSS RAGLQFPVGR VHRLLRKGNY AERVGAGAPV YLAAVLEYLT AEILELAGNA ARDNKKTRII PRHLQLAVR NDEELNKLLG RVTIAQGGVL PNIQSVLLPK KTESSKSAKS K UniProtKB: Histone H2A type 1 |
-Macromolecule #5: Histone H2B 1.1
| Macromolecule | Name: Histone H2B 1.1 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.655948 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKSAPAPKK GSKKAVTKTQ KKDGKKRRKT RKESYAIYVY KVLKQVHPDT GISSKAMSIM NSFVNDVFER IAGEASRLAH YNKRSTITS REIQTAVRLL LPGELAKHAV SEGTKAVTKY TSAK UniProtKB: Histone H2B 1.1 |
-Macromolecule #6: DNA (149-MER) Widom 601 sequence
| Macromolecule | Name: DNA (149-MER) Widom 601 sequence / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 49.175336 KDa |
| Sequence | String: (DT)(DC)(DT)(DA)(DG)(DG)(DT)(DG)(DA)(DC) (DC)(DA)(DT)(DC)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG) (DA)(DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA) (DT)(DT)(DG)(DG)(DT)(DC) ...String: (DT)(DC)(DT)(DA)(DG)(DG)(DT)(DG)(DA)(DC) (DC)(DA)(DT)(DC)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG) (DA)(DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT) (DA)(DG)(DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT) (DA)(DG) (DC)(DA)(DC)(DC)(DG)(DC)(DT) (DT)(DA)(DA)(DA)(DC)(DG)(DC)(DA)(DC)(DG) (DT)(DA)(DC) (DG)(DC)(DG)(DC)(DT)(DG) (DT)(DC)(DC)(DC)(DC)(DC)(DG)(DC)(DG)(DT) (DT)(DT)(DT)(DA) (DA)(DC)(DC)(DG)(DC) (DC)(DA)(DA)(DG)(DG)(DG)(DG)(DA)(DT)(DT) (DA)(DC)(DT)(DC)(DC) (DC)(DT)(DA)(DG) (DT)(DC)(DT)(DC)(DC)(DA)(DG)(DG)(DC)(DA) (DC)(DG)(DT)(DG)(DT)(DC) (DA)(DG)(DA) (DT)(DA)(DT)(DA)(DT)(DA)(DC)(DA)(DT)(DC) (DG)(DA)(DT)(DA)(DG)(DG)(DC) |
-Macromolecule #7: DNA (149-MER) Widom 601 sequence
| Macromolecule | Name: DNA (149-MER) Widom 601 sequence / type: dna / ID: 7 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 49.606586 KDa |
| Sequence | String: (DG)(DC)(DC)(DT)(DA)(DT)(DC)(DG)(DA)(DT) (DG)(DT)(DA)(DT)(DA)(DT)(DA)(DT)(DC)(DT) (DG)(DA)(DC)(DA)(DC)(DG)(DT)(DG)(DC) (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DC)(DT)(DA) (DG) (DG)(DG)(DA)(DG)(DT)(DA) ...String: (DG)(DC)(DC)(DT)(DA)(DT)(DC)(DG)(DA)(DT) (DG)(DT)(DA)(DT)(DA)(DT)(DA)(DT)(DC)(DT) (DG)(DA)(DC)(DA)(DC)(DG)(DT)(DG)(DC) (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DC)(DT)(DA) (DG) (DG)(DG)(DA)(DG)(DT)(DA)(DA)(DT) (DC)(DC)(DC)(DC)(DT)(DT)(DG)(DG)(DC)(DG) (DG)(DT) (DT)(DA)(DA)(DA)(DA)(DC)(DG) (DC)(DG)(DG)(DG)(DG)(DG)(DA)(DC)(DA)(DG) (DC)(DG)(DC) (DG)(DT)(DA)(DC)(DG)(DT) (DG)(DC)(DG)(DT)(DT)(DT)(DA)(DA)(DG)(DC) (DG)(DG)(DT)(DG) (DC)(DT)(DA)(DG)(DA) (DG)(DC)(DT)(DG)(DT)(DC)(DT)(DA)(DC)(DG) (DA)(DC)(DC)(DA)(DA) (DT)(DT)(DG)(DA) (DG)(DC)(DG)(DG)(DC)(DC)(DT)(DC)(DG)(DG) (DC)(DA)(DC)(DC)(DG)(DG) (DG)(DA)(DT) (DT)(DC)(DT)(DG)(DA)(DT)(DG)(DG)(DT)(DC) (DA)(DC)(DC)(DT)(DA)(DG)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 2 / Number real images: 22155 / Average electron dose: 37.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)