[English] 日本語
 Yorodumi
Yorodumi- EMDB-15718: Open state GluA1/A2 AMPA receptor in complex with TARP gamma 8 an... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Open state GluA1/A2 AMPA receptor in complex with TARP gamma 8 and ligand JNJ-61432059 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | AMPAR / ion channels / neurotransmission / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationPhase 0 - rapid depolarisation / Phase 2 - plateau phase / Cargo concentration in the ER / axonal spine / positive regulation of locomotion involved in locomotory behavior / positive regulation of membrane potential / COPII-mediated vesicle transport / cellular response to ammonium ion / response to sucrose / L-type voltage-gated calcium channel complex ...Phase 0 - rapid depolarisation / Phase 2 - plateau phase / Cargo concentration in the ER / axonal spine / positive regulation of locomotion involved in locomotory behavior / positive regulation of membrane potential / COPII-mediated vesicle transport / cellular response to ammonium ion / response to sucrose / L-type voltage-gated calcium channel complex / neuron spine / myosin V binding / postsynaptic neurotransmitter receptor diffusion trapping / proximal dendrite / regulation of AMPA receptor activity / regulation of monoatomic ion transmembrane transport / Trafficking of AMPA receptors / channel regulator activity / LGI-ADAM interactions / protein phosphatase 2B binding / response to arsenic-containing substance / cellular response to L-glutamate / cellular response to dsRNA / dendritic spine membrane / long-term synaptic depression / beta-2 adrenergic receptor binding / Synaptic adhesion-like molecules / cellular response to peptide hormone stimulus / spine synapse / response to morphine / dendritic spine neck / neuronal cell body membrane / dendritic spine head / cellular response to amine stimulus / response to psychosocial stress / peptide hormone receptor binding / spinal cord development / Activation of AMPA receptors / perisynaptic space / ligand-gated monoatomic cation channel activity / protein kinase A binding / AMPA glutamate receptor activity / Trafficking of GluR2-containing AMPA receptors / transmission of nerve impulse / response to lithium ion / behavioral response to pain / kainate selective glutamate receptor activity / cellular response to glycine / AMPA glutamate receptor complex / adenylate cyclase binding / extracellularly glutamate-gated ion channel activity / immunoglobulin binding / asymmetric synapse / ionotropic glutamate receptor complex / conditioned place preference / response to electrical stimulus / regulation of receptor recycling / excitatory synapse / G-protein alpha-subunit binding / glutamate receptor binding / Unblocking of NMDA receptors, glutamate binding and activation / positive regulation of excitatory postsynaptic potential / positive regulation of synaptic transmission / long-term memory / positive regulation of synaptic transmission, glutamatergic / regulation of postsynaptic membrane neurotransmitter receptor levels / regulation of synaptic transmission, glutamatergic / postsynaptic density, intracellular component / neuronal action potential / response to fungicide / voltage-gated calcium channel activity / glutamate-gated receptor activity / synapse assembly / regulation of long-term synaptic depression / cytoskeletal protein binding / extracellular ligand-gated monoatomic ion channel activity / cellular response to brain-derived neurotrophic factor stimulus / glutamate-gated calcium ion channel activity / presynaptic active zone membrane / somatodendritic compartment / ionotropic glutamate receptor binding / dendrite membrane / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / dendrite cytoplasm / ionotropic glutamate receptor signaling pathway / synaptic membrane / dendritic shaft / SNARE binding / PDZ domain binding / response to cocaine / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / synaptic transmission, glutamatergic / calcium channel regulator activity / protein tetramerization / cellular response to amino acid stimulus / response to nutrient levels / neuromuscular junction / establishment of protein localization / recycling endosome / postsynaptic density membrane Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
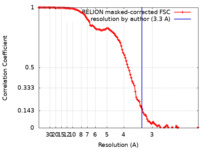
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||
 Authors Authors | Zhang D / Lape R / Shaikh S / Kohegyi B / Watson JF / Cais O / Nakagawa T / Greger IH | ||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Modulatory mechanisms of TARP γ8-selective AMPA receptor therapeutics. Authors: Danyang Zhang / Remigijus Lape / Saher A Shaikh / Bianka K Kohegyi / Jake F Watson / Ondrej Cais / Terunaga Nakagawa / Ingo H Greger /    Abstract: AMPA glutamate receptors (AMPARs) mediate excitatory neurotransmission throughout the brain. Their signalling is uniquely diversified by brain region-specific auxiliary subunits, providing an ...AMPA glutamate receptors (AMPARs) mediate excitatory neurotransmission throughout the brain. Their signalling is uniquely diversified by brain region-specific auxiliary subunits, providing an opportunity for the development of selective therapeutics. AMPARs associated with TARP γ8 are enriched in the hippocampus, and are targets of emerging anti-epileptic drugs. To understand their therapeutic activity, we determined cryo-EM structures of the GluA1/2-γ8 receptor associated with three potent, chemically diverse ligands. We find that despite sharing a lipid-exposed and water-accessible binding pocket, drug action is differentially affected by binding-site mutants. Together with patch-clamp recordings and MD simulations we also demonstrate that ligand-triggered reorganisation of the AMPAR-TARP interface contributes to modulation. Unexpectedly, one ligand (JNJ-61432059) acts bifunctionally, negatively affecting GluA1 but exerting positive modulatory action on GluA2-containing AMPARs, in a TARP stoichiometry-dependent manner. These results further illuminate the action of TARPs, demonstrate the sensitive balance between positive and negative modulatory action, and provide a mechanistic platform for development of both positive and negative selective AMPAR modulators. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15718.map.gz emd_15718.map.gz | 12.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15718-v30.xml emd-15718-v30.xml emd-15718.xml emd-15718.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15718_fsc.xml emd_15718_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_15718.png emd_15718.png | 129.4 KB | ||
| Filedesc metadata |  emd-15718.cif.gz emd-15718.cif.gz | 7.1 KB | ||
| Others |  emd_15718_additional_1.map.gz emd_15718_additional_1.map.gz emd_15718_half_map_1.map.gz emd_15718_half_map_1.map.gz emd_15718_half_map_2.map.gz emd_15718_half_map_2.map.gz | 4.9 MB 97 MB 97 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15718 http://ftp.pdbj.org/pub/emdb/structures/EMD-15718 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15718 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15718 | HTTPS FTP |
-Related structure data
| Related structure data |  8ayoMC  8aylC  8aymC  8aynC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15718.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15718.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
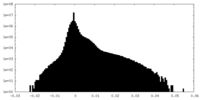
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_15718_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
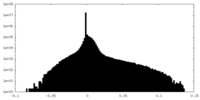
| Density Histograms |
-Half map: #1
| File | emd_15718_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
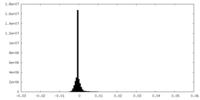
| Density Histograms |
-Half map: #2
| File | emd_15718_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
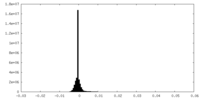
| Density Histograms |
- Sample components
Sample components
+Entire : GluA1/A2 AMPA receptor in complex with TARP gamma 8 and ligand JN...
+Supramolecule #1: GluA1/A2 AMPA receptor in complex with TARP gamma 8 and ligand JN...
+Macromolecule #1: Isoform Flip of Glutamate receptor 1
+Macromolecule #2: Isoform Flip of Glutamate receptor 2
+Macromolecule #3: Voltage-dependent calcium channel gamma-8 subunit
+Macromolecule #4: GLUTAMIC ACID
+Macromolecule #5: CYCLOTHIAZIDE
+Macromolecule #6: (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate
+Macromolecule #7: PALMITIC ACID
+Macromolecule #8: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
+Macromolecule #9: 5-[2-(4-fluorophenyl)-7-(4-oxidanylpiperidin-1-yl)pyrazolo[1,5-c]...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)







































 Homo sapiens (human)
Homo sapiens (human)