+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ASCC3 in complex with ASC1 | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | helicase / zinc finger / activator / DNA binding / DNA BINDING PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationALKBH3 mediated reversal of alkylation damage / RQC-trigger complex / regulation of myoblast differentiation / DNA alkylation repair / ribosome disassembly / DNA repair complex / ribosome-associated ubiquitin-dependent protein catabolic process / DNA 3'-5' helicase / 3'-5' DNA helicase activity / histone acetyltransferase binding ...ALKBH3 mediated reversal of alkylation damage / RQC-trigger complex / regulation of myoblast differentiation / DNA alkylation repair / ribosome disassembly / DNA repair complex / ribosome-associated ubiquitin-dependent protein catabolic process / DNA 3'-5' helicase / 3'-5' DNA helicase activity / histone acetyltransferase binding / ubiquitin-like protein ligase binding / estrogen receptor signaling pathway / rescue of stalled cytosolic ribosome / cytosolic ribosome / nuclear estrogen receptor binding / nuclear receptor binding / neuromuscular junction / protease binding / transcription coactivator activity / nuclear speck / nuclear body / centrosome / regulation of DNA-templated transcription / protein kinase binding / positive regulation of DNA-templated transcription / ATP hydrolysis activity / protein-containing complex / RNA binding / zinc ion binding / nucleoplasm / ATP binding / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
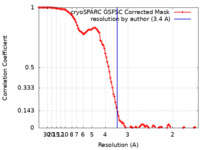
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||||||||
 Authors Authors | Jia J / Hilal T / Loll B / Wahl MC | ||||||||||||||||||
| Funding support |  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-EM structure of ASCC3 in complex with ASC1 Authors: Jia J / Hilal T / Loll B / Wahl MC | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15521.map.gz emd_15521.map.gz | 22.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15521-v30.xml emd-15521-v30.xml emd-15521.xml emd-15521.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15521_fsc.xml emd_15521_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15521.png emd_15521.png | 56.8 KB | ||
| Filedesc metadata |  emd-15521.cif.gz emd-15521.cif.gz | 7.9 KB | ||
| Others |  emd_15521_half_map_1.map.gz emd_15521_half_map_1.map.gz emd_15521_half_map_2.map.gz emd_15521_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15521 http://ftp.pdbj.org/pub/emdb/structures/EMD-15521 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15521 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15521 | HTTPS FTP |
-Related structure data
| Related structure data |  8alzMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15521.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15521.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
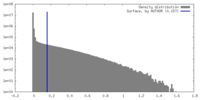
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_15521_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
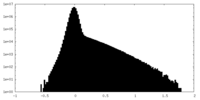
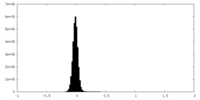
| Density Histograms |
-Half map: #1
| File | emd_15521_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
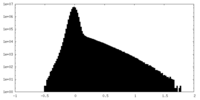
| Density Histograms |
- Sample components
Sample components
-Entire : ASCC3-ASC1
| Entire | Name: ASCC3-ASC1 |
|---|---|
| Components |
|
-Supramolecule #1: ASCC3-ASC1
| Supramolecule | Name: ASCC3-ASC1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Protein complex was produced in baculovirus-insect system. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 270 KDa |
-Macromolecule #1: Activating signal cointegrator 1
| Macromolecule | Name: Activating signal cointegrator 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 66.650656 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GAEFMAVAGA VSGEPLVHWC TQQLRKTFGL DVSEEIIQYV LSIESAEEIR EYVTDLLQGN EGKKGQFIEE LITKWQKNDQ ELISDPLQQ CFKKDEILDG QKSGDHLKRG RKKGRNRQEV PAFTEPDTTA EVKTPFDLAK AQENSNSVKK KTKFVNLYTR E GQDRLAVL ...String: GAEFMAVAGA VSGEPLVHWC TQQLRKTFGL DVSEEIIQYV LSIESAEEIR EYVTDLLQGN EGKKGQFIEE LITKWQKNDQ ELISDPLQQ CFKKDEILDG QKSGDHLKRG RKKGRNRQEV PAFTEPDTTA EVKTPFDLAK AQENSNSVKK KTKFVNLYTR E GQDRLAVL LPGRHPCDCL GQKHKLINNC LICGRIVCEQ EGSGPCLFCG TLVCTHEEQD ILQRDSNKSQ KLLKKLMSGV EN SGKVDIS TKDLLPHQEL RIKSGLEKAI KHKDKLLEFD RTSIRRTQVI DDESDYFASD SNQWLSKLER ETLQKREEEL REL RHASRL SKKVTIDFAG RKILEEENSL AEYHSRLDET IQAIANGTLN QPLTKLDRSS EEPLGVLVNP NMYQSPPQWV DHTG AASQK KAFRSSGFGL EFNSFQHQLR IQDQEFQEGF DGGWCLSVHQ PWASLLVRGI KRVEGRSWYT PHRGRLWIAA TAKKP SPQE VSELQATYRL LRGKDVEFPN DYPSGCLLGC VDLIDCLSQK QFKEQFPDIS QESDSPFVFI CKNPQEMVVK FPIKGN PKI WKLDSKIHQG AKKGLMKQNK AV UniProtKB: Activating signal cointegrator 1 |
-Macromolecule #2: Activating signal cointegrator 1 complex subunit 3
| Macromolecule | Name: Activating signal cointegrator 1 complex subunit 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 206.276094 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GAEFHYPHVY DSQAEAMKTS AFIAGAKMIL PEGIQRENNK LYEEVRIPYS EPMPLSFEEK PVYIQDLDEI GQLAFKGMKR LNRIQSIVF ETAYNTNENM LICAPTGAGK TNIAMLTVLH EIRQHFQQGV IKKNEFKIVY VAPMKALAAE MTDYFSRRLE P LGIIVKEL ...String: GAEFHYPHVY DSQAEAMKTS AFIAGAKMIL PEGIQRENNK LYEEVRIPYS EPMPLSFEEK PVYIQDLDEI GQLAFKGMKR LNRIQSIVF ETAYNTNENM LICAPTGAGK TNIAMLTVLH EIRQHFQQGV IKKNEFKIVY VAPMKALAAE MTDYFSRRLE P LGIIVKEL TGDMQLSKSE ILRTQMLVTT PEKWDVVTRK SVGDVALSQI VRLLILDEVH LLHEDRGPVL ESIVARTLRQ VE STQSMIR ILGLSATLPN YLDVATFLHV NPYIGLFFFD GRFRPVPLGQ TFLGIKCANK MQQLNNMDEV CYENVLKQVK AGH QVMVFV HARNATVRTA MSLIERAKNC GHIPFFFPTQ GHDYVLAEKQ VQRSRNKQVR ELFPDGFSIH HAGMLRQDRN LVEN LFSNG HIKVLVCTAT LAWGVNLPAH AVIIKGTQIY AAKRGSFVDL GILDVMQIFG RAGRPQFDKF GEGIIITTHD KLSHY LTLL TQRNPIESQF LESLADNLNA EIALGTVTNV EEAVKWISYT YLYVRMRANP LAYGISHKAY QIDPTLRKHR EQLVIE VGR KLDKAQMIRF EERTGYFSST DLGRTASHYY IKYNTIETFN ELFDAHKTEG DIFAIVSKAE EFDQIKVREE EIEELDT LL SNFCELSTPG GVENSYGKIN ILLQTYISRG EMDSFSLISD SAYVAQNAAR IVRALFEIAL RKRWPTMTYR LLNLSKVI D KRLWGWASPL RQFSILPPHI LTRLEEKKLT VDKLKDMRKD EIGHILHHVN IGLKVKQCVH QIPSVMMEAS IQPITRTVL RVTLSIYADF TWNDQVHGTV GEPWWIWVED PTNDHIYHSE YFLALKKQVI SKEAQLLVFT IPIFEPLPSQ YYIRAVSDRW LGAEAVCII NFQHLILPER HPPHTELLDL QPLPITALGC KAYEALYNFS HFNPVQTQIF HTLYHTDCNV LLGAPTGSGK T VAAELAIF RVFNKYPTSK AVYIAPLKAL VRERMDDWKV RIEEKLGKKV IELTGDVTPD MKSIAKADLI VTTPEKWDGV SR SWQNRNY VQQVTILIID EIHLLGEERG PVLEVIVSRT NFISSHTEKP VRIVGLSTAL ANARDLADWL NIKQMGLFNF RPS VRPVPL EVHIQGFPGQ HYCPRMASMN KPAFQAIRSH SPAKPVLIFV SSRRQTRLTA LELIAFLATE EDPKQWLNMD EREM ENIIA TVRDSNLKLT LAFGIGMHHA GLHERDRKTV EELFVNCKVQ VLIATSTLAW GVNFPAHLVI IKGTEYYDGK TRRYV DFPI TDVLQMMGRA GRPQFDDQGK AVILVHDIKK DFYKKFLYEP FPVESSLLGV LSDHLNAEIA GGTITSKQDA LDYITW TYF FRRLIMNPSY YNLGDVSHDS VNKFLSHLIE KSLIELELSY CIEIGEDNRS IEPLTYGRIA SYYYLKHQTV KMFKDRL KP ECSTEELLSI LSDAEEYTDL PVRHNEDHMN SELAKCLPIE SNPHSFDSPH TKAHLLLQAH LSRAMLPCPD YDTDTKTV L DQALRVCQAM LDVAANQGWL VTVLNITNLI QMVIQGRWLK DSSLLTLPNI ENHHLHLFKK WKPIMKGPHA RGRTSIESL PELIHACGGK DHVFSSMVES ELHAAKTKQA WNFLSHLPVI NVGISVKGSW DDLVEGHNEL SVSTLTADKR DDNKWIKLHA DQEYVLQVS LQRVHFGFHK GKPESCAVTP RFPKSKDEGW FLILGEVDKR ELIALKRVGY IRNHHVASLS FYTPEIPGRY I YTLYFMSD CYLGLDQQYD IYLNVTQASL SAQVNTKVSD SLTDLALK UniProtKB: Activating signal cointegrator 1 complex subunit 3 |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.15 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Solutions were made fresh from concentrated to avoid microbial contamination. | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Number real images: 6267 / Average exposure time: 40.57 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)