[English] 日本語
 Yorodumi
Yorodumi- EMDB-15516: Cryo-EM Snapshots of Nanodisc-Embedded Native Eukaryotic Membrane... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Snapshots of Nanodisc-Embedded Native Eukaryotic Membrane Proteins | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
| Biological species |  Thermochaetoides thermophila (fungus) Thermochaetoides thermophila (fungus) | |||||||||||||||||||||||||||
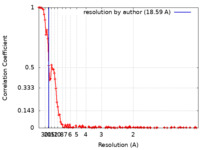
| Method | single particle reconstruction / cryo EM / Resolution: 18.59 Å | |||||||||||||||||||||||||||
 Authors Authors | Janson K / Kyrilis FL / Tueting C / Alfes M / Das M / Traeger TK / Schmidt C / Hamdi F / Keller S / Meister A / Kastritis PL | |||||||||||||||||||||||||||
| Funding support |  Germany, European Union, Germany, European Union,  France, France,  Austria, 8 items Austria, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Biomacromolecules / Year: 2022 Journal: Biomacromolecules / Year: 2022Title: Cryo-Electron Microscopy Snapshots of Eukaryotic Membrane Proteins in Native Lipid-Bilayer Nanodiscs. Authors: Kevin Janson / Fotis L Kyrilis / Christian Tüting / Marie Alfes / Manabendra Das / Toni K Träger / Carla Schmidt / Farzad Hamdi / Carolyn Vargas / Sandro Keller / Annette Meister / Panagiotis L Kastritis /   Abstract: New technologies for purifying membrane-bound protein complexes in combination with cryo-electron microscopy (EM) have recently allowed the exploration of such complexes under near-native conditions. ...New technologies for purifying membrane-bound protein complexes in combination with cryo-electron microscopy (EM) have recently allowed the exploration of such complexes under near-native conditions. In particular, polymer-encapsulated nanodiscs enable the study of membrane proteins at high resolution while retaining protein-protein and protein-lipid interactions within a lipid bilayer. However, this powerful technology has not been exploited to address the important question of how endogenous─as opposed to overexpressed─membrane proteins are organized within a lipid environment. In this work, we demonstrate that biochemical enrichment protocols for native membrane-protein complexes from in combination with polymer-based lipid-bilayer nanodiscs provide a substantial improvement in the quality of recovered endogenous membrane-protein complexes. Mass spectrometry results revealed ∼1123 proteins, while multiple 2D class averages and two 3D reconstructions from cryo-EM data furnished prominent structural signatures. This integrated methodological approach to enriching endogenous membrane-protein complexes provides unprecedented opportunities for a deeper understanding of eukaryotic membrane proteomes. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15516.map.gz emd_15516.map.gz | 153.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15516-v30.xml emd-15516-v30.xml emd-15516.xml emd-15516.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15516_fsc.xml emd_15516_fsc.xml | 16.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_15516.png emd_15516.png | 22.9 KB | ||
| Others |  emd_15516_half_map_1.map.gz emd_15516_half_map_1.map.gz emd_15516_half_map_2.map.gz emd_15516_half_map_2.map.gz | 151.8 MB 151.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15516 http://ftp.pdbj.org/pub/emdb/structures/EMD-15516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15516 | HTTPS FTP |
-Validation report
| Summary document |  emd_15516_validation.pdf.gz emd_15516_validation.pdf.gz | 836.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15516_full_validation.pdf.gz emd_15516_full_validation.pdf.gz | 835.6 KB | Display | |
| Data in XML |  emd_15516_validation.xml.gz emd_15516_validation.xml.gz | 20 KB | Display | |
| Data in CIF |  emd_15516_validation.cif.gz emd_15516_validation.cif.gz | 26.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15516 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15516 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15516 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15516 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15516.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15516.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.5918 Å | ||||||||||||||||||||||||||||||||||||
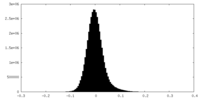
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_15516_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
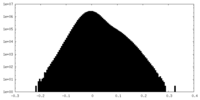
| Density Histograms |
-Half map: #1
| File | emd_15516_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SB-DIBMA solubilized Chaetomium thermophilum membranes mMW fraction
| Entire | Name: SB-DIBMA solubilized Chaetomium thermophilum membranes mMW fraction |
|---|---|
| Components |
|
-Supramolecule #1: SB-DIBMA solubilized Chaetomium thermophilum membranes mMW fraction
| Supramolecule | Name: SB-DIBMA solubilized Chaetomium thermophilum membranes mMW fraction type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 Details: Sample was created by solubilizing native chaetomium termophilum membranes with the aid of the copolymer SB-DIBMA |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) / Tissue: mycelia Thermochaetoides thermophila (fungus) / Tissue: mycelia |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: Solutions were freshly prepared, sterile filtrated, and sonicated before usage | |||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting time of 12 s Blotforce of 0. | |||||||||
| Details | This sample was purified by size exclusion chromatography. Subsequently, multiple fractions in the medium molecular weight region were pooled together to obtain a higher protein concentration. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Temperature | Min: 77.15 K / Max: 103.15 K |
| Alignment procedure | Coma free - Residual tilt: 14.7 mrad |
| Details | Grid screening was performed manually until criteria for good acquisition areas was narrowed down. |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4048 pixel / Digitization - Dimensions - Height: 4048 pixel / Number grids imaged: 1 / Number real images: 5912 / Average electron dose: 64.72 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 236566 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 240000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)