+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vaccinia C16 N-terminal domains | |||||||||||||||||||||
 Map data Map data | Map of C16 N-terminal domains in the C16-Ku70/Ku80 complex | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | C16 vaccinia virus protein Ku70/Ku80 DNA binding inhibition / VIRAL PROTEIN | |||||||||||||||||||||
| Function / homology | Poxvirus C4/C10 / Poxvirus C4/C10 protein / Protein C10 Function and homology information Function and homology information | |||||||||||||||||||||
| Biological species |  Vaccinia virus / Vaccinia virus /  Vaccinia virus Western Reserve Vaccinia virus Western Reserve | |||||||||||||||||||||
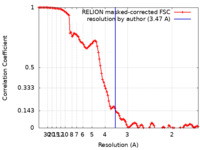
| Method | single particle reconstruction / cryo EM / Resolution: 3.47 Å | |||||||||||||||||||||
 Authors Authors | Rivera-Calzada A / Arribas-Bosacoma R / Pearl LH / Llorca O | |||||||||||||||||||||
| Funding support |  Spain, Spain,  United Kingdom, 6 items United Kingdom, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus. Authors: Angel Rivera-Calzada / Raquel Arribas-Bosacoma / Alba Ruiz-Ramos / Paloma Escudero-Bravo / Jasminka Boskovic / Rafael Fernandez-Leiro / Antony W Oliver / Laurence H Pearl / Oscar Llorca /   Abstract: Detection of cytosolic DNA is a central element of the innate immunity system against viral infection. The Ku heterodimer, a component of the NHEJ pathway of DNA repair in the nucleus, functions as ...Detection of cytosolic DNA is a central element of the innate immunity system against viral infection. The Ku heterodimer, a component of the NHEJ pathway of DNA repair in the nucleus, functions as DNA sensor that detects dsDNA of viruses that replicate in the cytoplasm. Vaccinia virus expresses two proteins, C4 and C16, that inactivate DNA sensing and enhance virulence. The structural basis for this is unknown. Here we determine the structure of the C16 - Ku complex using cryoEM. Ku binds dsDNA by a preformed ring but C16 sterically blocks this access route, abrogating binding to a dsDNA end and its insertion into DNA-PK, thereby averting signalling into the downstream innate immunity system. C4 replicates these activities using a domain with 54% identity to C16. Our results reveal how vaccinia virus subverts the capacity of Ku to recognize viral DNA. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15414.map.gz emd_15414.map.gz | 90.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15414-v30.xml emd-15414-v30.xml emd-15414.xml emd-15414.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15414_fsc.xml emd_15414_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_15414.png emd_15414.png | 67.8 KB | ||
| Filedesc metadata |  emd-15414.cif.gz emd-15414.cif.gz | 6.2 KB | ||
| Others |  emd_15414_half_map_1.map.gz emd_15414_half_map_1.map.gz emd_15414_half_map_2.map.gz emd_15414_half_map_2.map.gz | 84.2 MB 84.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15414 http://ftp.pdbj.org/pub/emdb/structures/EMD-15414 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15414 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15414 | HTTPS FTP |
-Related structure data
| Related structure data |  8ag3MC  8ag4C  8ag5C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15414.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15414.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of C16 N-terminal domains in the C16-Ku70/Ku80 complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
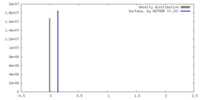
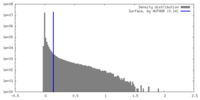
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 1
| File | emd_15414_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
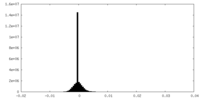
| Density Histograms |
-Half map: Half map 2
| File | emd_15414_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
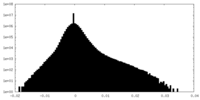
| Density Histograms |
- Sample components
Sample components
-Entire : C16 N-terminal domains in the C16-Ku70/Ku80 complex
| Entire | Name: C16 N-terminal domains in the C16-Ku70/Ku80 complex |
|---|---|
| Components |
|
-Supramolecule #1: C16 N-terminal domains in the C16-Ku70/Ku80 complex
| Supramolecule | Name: C16 N-terminal domains in the C16-Ku70/Ku80 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Vaccinia virus / Strain: Western Reserve Vaccinia virus / Strain: Western Reserve |
| Molecular weight | Theoretical: 230 KDa |
-Macromolecule #1: Protein C10
| Macromolecule | Name: Protein C10 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vaccinia virus Western Reserve / Strain: Western Reserve Vaccinia virus Western Reserve / Strain: Western Reserve |
| Molecular weight | Theoretical: 42.47668 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDIYDDKGLQ TIKLFNNEFD CIRNDIRELF KHVTDSDSIQ LPMEDNSDII ENIRKILYRR LKNVECVDID STITFMKYDP NDDNKRTCS NWVPLTNNYM EYCLVIYLET PICGGKIKLY HPTGNIKSDK DIMFAKTLDF KSKKVLTGRK TIAVLDISVS Y NRSMTTIH ...String: MDIYDDKGLQ TIKLFNNEFD CIRNDIRELF KHVTDSDSIQ LPMEDNSDII ENIRKILYRR LKNVECVDID STITFMKYDP NDDNKRTCS NWVPLTNNYM EYCLVIYLET PICGGKIKLY HPTGNIKSDK DIMFAKTLDF KSKKVLTGRK TIAVLDISVS Y NRSMTTIH YNDDVDIDIH TDKNGKELCY CYITIDDHYL VDVETIGVIV NRSGKCLLVN NHLGIGIVKD KRISDSFGDV CM DTIFDFS EARELFSLTN DDNRNIAWDT DKLDDDTDIW TPVTEDDYKF LSRLVLYAKS QSDTVFDYYV LTGDTEPPTV FIF KVTRFY FNMPKGGENL YFQGWSHPQF EKGGGSGGGS GGSSAWSHPQ FEK UniProtKB: Protein C10 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.9 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Mesh: 300 | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 13216 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Initial model for chains C and D was generated using AlphaFold2 |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8ag3: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)