+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | C1 Cyanobacteria sp PCC 7001 RuBisCO | |||||||||
 Map data Map data | Cryo-EM structure of an a-carboxysome RuBisCO enzyme with C1 symmetry at 3.8 A resolution | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationphotorespiration / carboxysome / ribulose-bisphosphate carboxylase / ribulose-bisphosphate carboxylase activity / reductive pentose-phosphate cycle / monooxygenase activity / magnesium ion binding Similarity search - Function | |||||||||
| Biological species |  Cyanobium sp. PCC 7001 (bacteria) Cyanobium sp. PCC 7001 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.79 Å | |||||||||
 Authors Authors | Evans SL / Mann D / Bergeron JRC | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Single-particle cryo-EM analysis of the shell architecture and internal organization of an intact a-carboxysome Authors: Evans SL / Al-Hazeem MJ / Mann D / Smetacek N / Beavil AJ / Sun Y / Chen T / Dykes GF / Liu L / Bergeron JRC | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15409.map.gz emd_15409.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15409-v30.xml emd-15409-v30.xml emd-15409.xml emd-15409.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15409_fsc.xml emd_15409_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_15409.png emd_15409.png | 59.1 KB | ||
| Others |  emd_15409_half_map_1.map.gz emd_15409_half_map_1.map.gz emd_15409_half_map_2.map.gz emd_15409_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15409 http://ftp.pdbj.org/pub/emdb/structures/EMD-15409 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15409 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15409 | HTTPS FTP |
-Validation report
| Summary document |  emd_15409_validation.pdf.gz emd_15409_validation.pdf.gz | 903.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15409_full_validation.pdf.gz emd_15409_full_validation.pdf.gz | 903.2 KB | Display | |
| Data in XML |  emd_15409_validation.xml.gz emd_15409_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  emd_15409_validation.cif.gz emd_15409_validation.cif.gz | 21.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15409 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15409 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15409 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15409 | HTTPS FTP |
-Related structure data
| Related structure data |  8cmyM  8aft M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15409.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15409.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of an a-carboxysome RuBisCO enzyme with C1 symmetry at 3.8 A resolution | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.11 Å | ||||||||||||||||||||||||||||||||||||
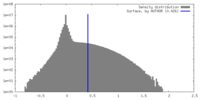
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_15409_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
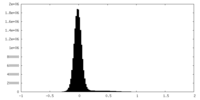
| Density Histograms |
-Half map: #1
| File | emd_15409_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RuBisCO
| Entire | Name: RuBisCO |
|---|---|
| Components |
|
-Supramolecule #1: RuBisCO
| Supramolecule | Name: RuBisCO / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Cyanobium sp. PCC 7001 (bacteria) Cyanobium sp. PCC 7001 (bacteria) |
-Macromolecule #1: Ribulose bisphosphate carboxylase large chain
| Macromolecule | Name: Ribulose bisphosphate carboxylase large chain / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO / EC number: ribulose-bisphosphate carboxylase |
|---|---|
| Source (natural) | Organism:  Cyanobium sp. PCC 7001 (bacteria) Cyanobium sp. PCC 7001 (bacteria) |
| Molecular weight | Theoretical: 52.526531 KDa |
| Sequence | String: MSKKYDAGVK EYRDTYWTPD YVPLDTDLLA CFKCTGQEGV PKEEVAAAVA AESSTGTWST VWSELLVDLD FYKGRCYRIE DVPGDKEAF YAFIAYPLDL FEEGSVTNVL TSLVGNVFGF KALRHLRLED IRFPMAFIKT CPGPPNGICV ERDRMNKYGR P LLGCTIKP ...String: MSKKYDAGVK EYRDTYWTPD YVPLDTDLLA CFKCTGQEGV PKEEVAAAVA AESSTGTWST VWSELLVDLD FYKGRCYRIE DVPGDKEAF YAFIAYPLDL FEEGSVTNVL TSLVGNVFGF KALRHLRLED IRFPMAFIKT CPGPPNGICV ERDRMNKYGR P LLGCTIKP KLGLSGKNYG RVVYECLRGG LDFTKDDENI NSQPFQRWQN RFEFVAEAVA LAQQETGEKK GHYLNCTAAT PE EMYERAE FAKELGQPII MHDYITGGFT ANTGLSKWCR KNGMLLHIHR AMHAVIDRHP KHGIHFRVLA KCLRLSGGDQ LHT GTVVGK LEGDRQTTLG FIDQLRESFI PEDRSRGNFF DQDWGSMPGV FAVASGGIHV WHMPALVAIF GDDSVLQFGG GTHG HPWGS AAGAAANRVA LEACVKARNA GREIEKESRD ILMEAAKHSP ELAIALETWK EIKFEFDTVD KLDVQ |
-Macromolecule #2: Ribulose bisphosphate carboxylase, small subunit
| Macromolecule | Name: Ribulose bisphosphate carboxylase, small subunit / type: protein_or_peptide / ID: 2 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Cyanobium sp. PCC 7001 (bacteria) Cyanobium sp. PCC 7001 (bacteria) |
| Molecular weight | Theoretical: 12.880533 KDa |
| Sequence | String: MPFKTVGDYQ TVATLETFGF LPPMTQDEIY DQIAYIIAQG WSPLIEHVHP SRSMATYWSY WKLPFFGEKD LGVIVSELEA CHRAYPDHH VRLVGYDAYT QSQGACFVVF EGR |
-Macromolecule #3: 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE
| Macromolecule | Name: 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 7 / Formula: CAP |
|---|---|
| Molecular weight | Theoretical: 356.115 Da |
| Chemical component information |  ChemComp-CAP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 8 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)