+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | FEN1 holoenzyme with a nicked DNA substrate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA / Replication / Complex / Ligase / PCNA / Ligation / FEN1 / Flap Endonuclease I / Okazaki fragment maturation | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
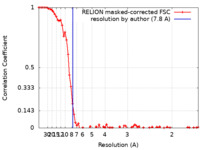
| Method | single particle reconstruction / cryo EM / Resolution: 7.8 Å | |||||||||
 Authors Authors | Blair K / Tehseen M / Raducanu VS / Shahid T / Lancey C / Cruehet R / Hamdan S / De Biasio A | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Mechanism of human Lig1 regulation by PCNA in Okazaki fragment sealing. Authors: Kerry Blair / Muhammad Tehseen / Vlad-Stefan Raducanu / Taha Shahid / Claudia Lancey / Fahad Rashid / Ramon Crehuet / Samir M Hamdan / Alfredo De Biasio /    Abstract: During lagging strand synthesis, DNA Ligase 1 (Lig1) cooperates with the sliding clamp PCNA to seal the nicks between Okazaki fragments generated by Pol δ and Flap endonuclease 1 (FEN1). We present ...During lagging strand synthesis, DNA Ligase 1 (Lig1) cooperates with the sliding clamp PCNA to seal the nicks between Okazaki fragments generated by Pol δ and Flap endonuclease 1 (FEN1). We present several cryo-EM structures combined with functional assays, showing that human Lig1 recruits PCNA to nicked DNA using two PCNA-interacting motifs (PIPs) located at its disordered N-terminus (PIP) and DNA binding domain (PIP). Once Lig1 and PCNA assemble as two-stack rings encircling DNA, PIP is released from PCNA and only PIP is required for ligation to facilitate the substrate handoff from FEN1. Consistently, we observed that PCNA forms a defined complex with FEN1 and nicked DNA, and it recruits Lig1 to an unoccupied monomer creating a toolbelt that drives the transfer of DNA to Lig1. Collectively, our results provide a structural model on how PCNA regulates FEN1 and Lig1 during Okazaki fragments maturation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15385.map.gz emd_15385.map.gz | 35.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15385-v30.xml emd-15385-v30.xml emd-15385.xml emd-15385.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15385_fsc.xml emd_15385_fsc.xml | 7.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_15385.png emd_15385.png | 59.6 KB | ||
| Filedesc metadata |  emd-15385.cif.gz emd-15385.cif.gz | 6.1 KB | ||
| Others |  emd_15385_half_map_1.map.gz emd_15385_half_map_1.map.gz emd_15385_half_map_2.map.gz emd_15385_half_map_2.map.gz | 29.5 MB 29.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15385 http://ftp.pdbj.org/pub/emdb/structures/EMD-15385 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15385 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15385 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15385.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15385.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_15385_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
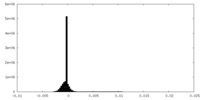
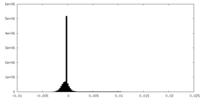
| Density Histograms |
-Half map: #1
| File | emd_15385_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
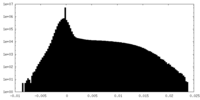
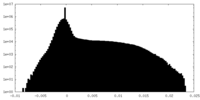
| Density Histograms |
- Sample components
Sample components
+Entire : complex of FEN1 with PCNA and DNA
+Supramolecule #1: complex of FEN1 with PCNA and DNA
+Supramolecule #2: Proliferating cell nuclear antigen and Flap Endonuclease I
+Supramolecule #3: Oligo19ddC, Oligo13P and Oligo32
+Macromolecule #1: Proliferating cell nuclear antigen
+Macromolecule #2: Proliferating cell nuclear antigen
+Macromolecule #3: Proliferating cell nuclear antigen
+Macromolecule #4: Flap Endonuclease I
+Macromolecule #5: Oligo19ddC
+Macromolecule #6: Oligo13P
+Macromolecule #7: Oligo32
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 300 sec. Details: The grid was coated with graphene oxide prior to use. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 77.0 K / Max: 77.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Average exposure time: 2.0 sec. / Average electron dose: 18.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller










 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)