[English] 日本語
 Yorodumi
Yorodumi- EMDB-15360: Structure of dimeric yeast RNA polymerase II bound to a transcrip... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of dimeric yeast RNA polymerase II bound to a transcription bubble (focused map of monomer 2) | |||||||||
 Map data Map data | monomer 2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription regulation / RNA 3' end processing / transcription termination / GENE REGULATION | |||||||||
| Biological species |  | |||||||||
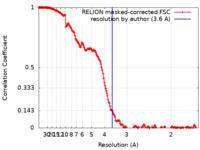
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Carminati M / Manav MC / Bellini D / Passmore LA | |||||||||
| Funding support | European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: A direct interaction between CPF and RNA Pol II links RNA 3' end processing to transcription. Authors: Manuel Carminati / Juan B Rodríguez-Molina / M Cemre Manav / Dom Bellini / Lori A Passmore /  Abstract: Transcription termination by RNA polymerase II (RNA Pol II) is linked to RNA 3' end processing by the cleavage and polyadenylation factor (CPF or CPSF). CPF contains endonuclease, poly(A) polymerase, ...Transcription termination by RNA polymerase II (RNA Pol II) is linked to RNA 3' end processing by the cleavage and polyadenylation factor (CPF or CPSF). CPF contains endonuclease, poly(A) polymerase, and protein phosphatase activities, which cleave and polyadenylate pre-mRNAs and dephosphorylate RNA Pol II to control transcription. Exactly how the RNA 3' end processing machinery is coupled to transcription remains unclear. Here, we combine in vitro reconstitution, structural studies, and genome-wide analyses to show that yeast CPF physically and functionally interacts with RNA Pol II. Surprisingly, CPF-mediated dephosphorylation promotes the formation of an RNA Pol II stalk-to-stalk homodimer in vitro. This dimer is compatible with transcription but not with the binding of transcription elongation factors. Disruption of the dimerization interface in cells causes transcription defects, including altered RNA Pol II abundance on protein-coding genes, tRNA genes, and intergenic regions. We hypothesize that RNA Pol II dimerization may provide a mechanistic basis for the allosteric model of transcription termination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15360.map.gz emd_15360.map.gz | 427.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15360-v30.xml emd-15360-v30.xml emd-15360.xml emd-15360.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15360_fsc.xml emd_15360_fsc.xml | 17.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_15360.png emd_15360.png | 110.6 KB | ||
| Filedesc metadata |  emd-15360.cif.gz emd-15360.cif.gz | 5 KB | ||
| Others |  emd_15360_half_map_1.map.gz emd_15360_half_map_1.map.gz emd_15360_half_map_2.map.gz emd_15360_half_map_2.map.gz | 380.5 MB 380.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15360 http://ftp.pdbj.org/pub/emdb/structures/EMD-15360 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15360 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15360 | HTTPS FTP |
-Validation report
| Summary document |  emd_15360_validation.pdf.gz emd_15360_validation.pdf.gz | 872.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15360_full_validation.pdf.gz emd_15360_full_validation.pdf.gz | 872.1 KB | Display | |
| Data in XML |  emd_15360_validation.xml.gz emd_15360_validation.xml.gz | 24.8 KB | Display | |
| Data in CIF |  emd_15360_validation.cif.gz emd_15360_validation.cif.gz | 32.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15360 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15360 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15360 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15360 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15360.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15360.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | monomer 2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: monomer 2
| File | emd_15360_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | monomer 2 | ||||||||||||
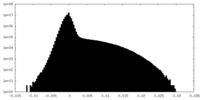
| Projections & Slices |
| ||||||||||||
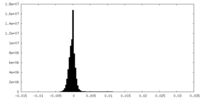
| Density Histograms |
-Half map: monomer 2
| File | emd_15360_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | monomer 2 | ||||||||||||
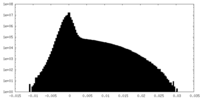
| Projections & Slices |
| ||||||||||||
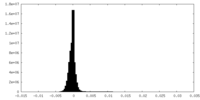
| Density Histograms |
- Sample components
Sample components
-Entire : S. cerevisiae RNA polymerase II
| Entire | Name: S. cerevisiae RNA polymerase II |
|---|---|
| Components |
|
-Supramolecule #1: S. cerevisiae RNA polymerase II
| Supramolecule | Name: S. cerevisiae RNA polymerase II / type: complex / ID: 1 / Parent: 0 Details: RNA polymerase 'stalk-to-stalk' homodimer loaded with a DNA-RNA scaffold mimicking a transcription bubble |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 552 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.1 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 0.005 % v/v Tween-20 was added to the sample just before vitrification to prevent preferred orientation problems. | ||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 45 sec. | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 4 seconds (force -12) before plunging. | ||||||||
| Details | The vitrified sample contained Pol II (with transcription bubble) bound to the CPF subcomplex Ref2:Glc7:Swd2. We observed a dimeric Pol II population (~10 % of particles) which is reported in the present deposition. |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Specialist optics | Energy filter - Slit width: 20 eV |
| Details | The movies were collected without tilt or with a tilt angle over a 33-45 degrees range. |
| Image recording | Image recording ID: 1 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 6 / Number real images: 25411 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Details | The movies were collected without tilt or with a tilt angle over a 30-40 degrees range. |
| Image recording | Image recording ID: 2 / Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Number real images: 2035 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 75000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Pol II structure from PDB: 5C4X was rigid fit into the dimeric Pol II EM density |
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)