+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Consensus reconstruction of the dextran utilisation system | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Membrane protein transporter / glycan transporter / SusCD / utilisome / TonB dependent transporter / TBDT / TRANSPORT PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) | ||||||||||||
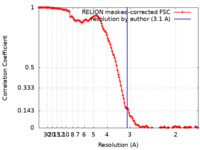
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||
 Authors Authors | White JBR / Silale A / Ranson NA / van den Berg B | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Outer membrane utilisomes mediate glycan uptake in gut Bacteroidetes. Authors: Joshua B R White / Augustinas Silale / Matthew Feasey / Tiaan Heunis / Yiling Zhu / Hong Zheng / Akshada Gajbhiye / Susan Firbank / Arnaud Baslé / Matthias Trost / David N Bolam / Bert van ...Authors: Joshua B R White / Augustinas Silale / Matthew Feasey / Tiaan Heunis / Yiling Zhu / Hong Zheng / Akshada Gajbhiye / Susan Firbank / Arnaud Baslé / Matthias Trost / David N Bolam / Bert van den Berg / Neil A Ranson /  Abstract: Bacteroidetes are abundant members of the human microbiota, utilizing a myriad of diet- and host-derived glycans in the distal gut. Glycan uptake across the bacterial outer membrane of these bacteria ...Bacteroidetes are abundant members of the human microbiota, utilizing a myriad of diet- and host-derived glycans in the distal gut. Glycan uptake across the bacterial outer membrane of these bacteria is mediated by SusCD protein complexes, comprising a membrane-embedded barrel and a lipoprotein lid, which is thought to open and close to facilitate substrate binding and transport. However, surface-exposed glycan-binding proteins and glycoside hydrolases also play critical roles in the capture, processing and transport of large glycan chains. The interactions between these components in the outer membrane are poorly understood, despite being crucial for nutrient acquisition by our colonic microbiota. Here we show that for both the levan and dextran utilization systems of Bacteroides thetaiotaomicron, the additional outer membrane components assemble on the core SusCD transporter, forming stable glycan-utilizing machines that we term utilisomes. Single-particle cryogenic electron microscopy structures in the absence and presence of substrate reveal concerted conformational changes that demonstrate the mechanism of substrate capture, and rationalize the role of each component in the utilisome. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15293.map.gz emd_15293.map.gz | 13.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15293-v30.xml emd-15293-v30.xml emd-15293.xml emd-15293.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15293_fsc.xml emd_15293_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_15293.png emd_15293.png | 67.8 KB | ||
| Masks |  emd_15293_msk_1.map emd_15293_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15293.cif.gz emd-15293.cif.gz | 6.1 KB | ||
| Others |  emd_15293_additional_1.map.gz emd_15293_additional_1.map.gz emd_15293_half_map_1.map.gz emd_15293_half_map_1.map.gz emd_15293_half_map_2.map.gz emd_15293_half_map_2.map.gz | 139.9 MB 140.5 MB 140.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15293 http://ftp.pdbj.org/pub/emdb/structures/EMD-15293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15293 | HTTPS FTP |
-Related structure data
| Related structure data |  8aa4MC  7znrC  7znsC  8a9yC  8aa0C  8aa1C  8aa2C  8aa3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15293.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15293.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
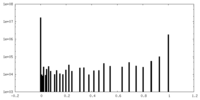
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
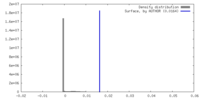
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15293_msk_1.map emd_15293_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
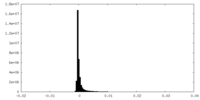
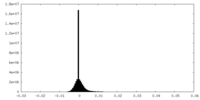
| Density Histograms |
-Additional map: #1
| File | emd_15293_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
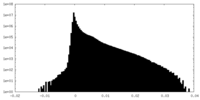
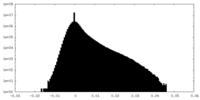
| Density Histograms |
-Half map: #1
| File | emd_15293_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
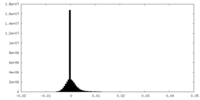
| Density Histograms |
-Half map: #2
| File | emd_15293_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
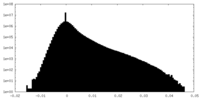
| Density Histograms |
- Sample components
Sample components
-Entire : Dextran utilisation machinery (utilisome)
| Entire | Name: Dextran utilisation machinery (utilisome) |
|---|---|
| Components |
|
-Supramolecule #1: Dextran utilisation machinery (utilisome)
| Supramolecule | Name: Dextran utilisation machinery (utilisome) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Sample for EM experiments was the purified dextran utilisome (Bt3088-Bt3090). Only SusC components were of sufficient resolution for model building and refinement. |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
-Macromolecule #1: SusC homolog
| Macromolecule | Name: SusC homolog / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria)Strain: ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50 |
| Molecular weight | Theoretical: 110.591242 KDa |
| Sequence | String: MEQSIKSKGF EHRLLLIMWG LLLSLSAFAQ QITVKGHVVD ATGEPVIGAS VIEGKSTNGT ITDIDGNFSL NVSANSALTI SFVGYKTQT VSVNGKTALK VTLQEDTEVL DEVVVVGYGT MKKSDLTGAV SSVGVKDIKD SPVANIGQAM QGKVSGVQII D AGKPGDNV ...String: MEQSIKSKGF EHRLLLIMWG LLLSLSAFAQ QITVKGHVVD ATGEPVIGAS VIEGKSTNGT ITDIDGNFSL NVSANSALTI SFVGYKTQT VSVNGKTALK VTLQEDTEVL DEVVVVGYGT MKKSDLTGAV SSVGVKDIKD SPVANIGQAM QGKVSGVQII D AGKPGDNV TIKIRGLGTI NNSNPLVVID GIPTDLGLSS LNMADVERVD VLKDASATAI YGSRGANGVV MITSKRGAEG AG KVTVNAN WAIQNATKVP DMLNAAQYAA LSNDMLSNND DNTNPYWADP SSLGKGTNWL DEMLRTGVKQ SYSVSYSGGT EKA HYYVSG GFLDQSGIVK SVNYRRFNFQ ANSDAQVNKW LKFTTNLTFS TDVKEGGTYS IGDAMKALPT QPVKNDDGSW SGPG QEAQW YGSIRNPIGT LHMMTNETKG YNFLANITGE ITFTKWLKLK STFGYDAKFW FADNFTPAYD WKPNPVEESS RYKSD NKSF TYLWDNYFVF DHTFAKKHRV GVMAGSSAQW NNYDYLNAQK NIFMFDNIHE MDNGEKMYSL GGSQSDWALL SLMARL NYS YEDKYLLTAT VRRDGSSRFG KNNRWGTFPS VSLAWRVSQE DWFPKDNFLM NDLKLRVGYG VTGNQEIGNY GFVASYN TG VYPFGNNNST ALVSTTLSNP NIHWEEVRQA NFGVDMSLFD SRVSLSLDAY IKNTNDMLVK ASIPITSGFE DTTETFTN A GKMRNKGVEM TLRTINLKGI FSWESALTAT YNKNEILDLN SETPMFINQI GNSYVTMLKA GYPINVFYGY VTDGLFQNW GEVNRHATQP GAAPGDIRFR DLNNDGVIND EDRTILGNPN PNWFFSLSNN LSYKGWELSV FLQGVAGNKI YNANNVDNEG MAAAYNQTT AVLNRWTGEG TSYSMPRAIW GDPNQNCRVS DRFVENGSYL RLKNITLSYT LPKKWLQKIQ LENARISFSC E NVATITRY SGFDPEVDVN GIDSSRYPIS RTFSMGLNFN F UniProtKB: SusC homolog |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 38.78 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)