[English] 日本語
 Yorodumi
Yorodumi- EMDB-15289: Levan utilisation machinery (utilisome) with levan fructo-oligosa... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Levan utilisation machinery (utilisome) with levan fructo-oligosaccharides DP 8-12 | ||||||||||||
 Map data Map data | Sharpened map from post-processing using an ad-hoc low-pass filter of 3.3 A | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Membrane protein transporter / glycan transporter / SusCD / utilisome / TonB dependent transporter / TBDT / TRANSPORT PROTEIN / levan | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationbeta-fructofuranosidase / sucrose alpha-glucosidase activity / cell outer membrane / carbohydrate metabolic process / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | White JBR / Silale A / Ranson NA / van den Berg B | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Outer membrane utilisomes mediate glycan uptake in gut Bacteroidetes. Authors: Joshua B R White / Augustinas Silale / Matthew Feasey / Tiaan Heunis / Yiling Zhu / Hong Zheng / Akshada Gajbhiye / Susan Firbank / Arnaud Baslé / Matthias Trost / David N Bolam / Bert van ...Authors: Joshua B R White / Augustinas Silale / Matthew Feasey / Tiaan Heunis / Yiling Zhu / Hong Zheng / Akshada Gajbhiye / Susan Firbank / Arnaud Baslé / Matthias Trost / David N Bolam / Bert van den Berg / Neil A Ranson /  Abstract: Bacteroidetes are abundant members of the human microbiota, utilizing a myriad of diet- and host-derived glycans in the distal gut. Glycan uptake across the bacterial outer membrane of these bacteria ...Bacteroidetes are abundant members of the human microbiota, utilizing a myriad of diet- and host-derived glycans in the distal gut. Glycan uptake across the bacterial outer membrane of these bacteria is mediated by SusCD protein complexes, comprising a membrane-embedded barrel and a lipoprotein lid, which is thought to open and close to facilitate substrate binding and transport. However, surface-exposed glycan-binding proteins and glycoside hydrolases also play critical roles in the capture, processing and transport of large glycan chains. The interactions between these components in the outer membrane are poorly understood, despite being crucial for nutrient acquisition by our colonic microbiota. Here we show that for both the levan and dextran utilization systems of Bacteroides thetaiotaomicron, the additional outer membrane components assemble on the core SusCD transporter, forming stable glycan-utilizing machines that we term utilisomes. Single-particle cryogenic electron microscopy structures in the absence and presence of substrate reveal concerted conformational changes that demonstrate the mechanism of substrate capture, and rationalize the role of each component in the utilisome. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15289.map.gz emd_15289.map.gz | 10.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15289-v30.xml emd-15289-v30.xml emd-15289.xml emd-15289.xml | 25.3 KB 25.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15289_fsc.xml emd_15289_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15289.png emd_15289.png | 69.7 KB | ||
| Masks |  emd_15289_msk_1.map emd_15289_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15289.cif.gz emd-15289.cif.gz | 7.9 KB | ||
| Others |  emd_15289_additional_1.map.gz emd_15289_additional_1.map.gz emd_15289_half_map_1.map.gz emd_15289_half_map_1.map.gz emd_15289_half_map_2.map.gz emd_15289_half_map_2.map.gz | 79.3 MB 79.5 MB 79.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15289 http://ftp.pdbj.org/pub/emdb/structures/EMD-15289 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15289 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15289 | HTTPS FTP |
-Related structure data
| Related structure data |  8aa0MC  7znrC  7znsC  8a9yC  8aa1C  8aa2C  8aa3C  8aa4C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15289.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15289.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map from post-processing using an ad-hoc low-pass filter of 3.3 A | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15289_msk_1.map emd_15289_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
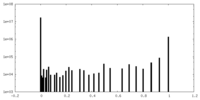
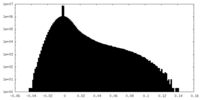
| Density Histograms |
-Additional map: Unsharpened map from 3D refinement
| File | emd_15289_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map from 3D refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
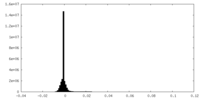
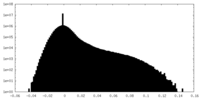
| Density Histograms |
-Half map: #2
| File | emd_15289_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
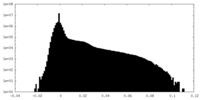
| Density Histograms |
-Half map: #1
| File | emd_15289_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
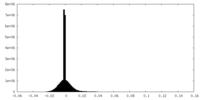
| Density Histograms |
- Sample components
Sample components
-Entire : Levan utilisation machinery (utilisome) with levan fructo-oligosa...
| Entire | Name: Levan utilisation machinery (utilisome) with levan fructo-oligosaccharides DP8-12 |
|---|---|
| Components |
|
-Supramolecule #1: Levan utilisation machinery (utilisome) with levan fructo-oligosa...
| Supramolecule | Name: Levan utilisation machinery (utilisome) with levan fructo-oligosaccharides DP8-12 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / Details: Levan utilisome comprised of Bt1760-Bt1763 |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
-Macromolecule #1: Glycoside hydrolase family 32
| Macromolecule | Name: Glycoside hydrolase family 32 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria)Strain: ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50 |
| Molecular weight | Theoretical: 59.299926 KDa |
| Recombinant expression | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
| Sequence | String: MMKNMILPIA FTALIASMTA CSDETDPILT QKNWDGTATY FQSSDEHGFS MYYKPQVGFV GDPMPFYDPV AKDFKVMYLQ DYRPNPEAT YHPIFGVATK DGATYESLGE LISCGGRDEQ DAAIGTGGTI YNPADKLYYT FYTGNKFKPS SDQNAQVVMV A TSPDFKTW ...String: MMKNMILPIA FTALIASMTA CSDETDPILT QKNWDGTATY FQSSDEHGFS MYYKPQVGFV GDPMPFYDPV AKDFKVMYLQ DYRPNPEAT YHPIFGVATK DGATYESLGE LISCGGRDEQ DAAIGTGGTI YNPADKLYYT FYTGNKFKPS SDQNAQVVMV A TSPDFKTW TKNRTFYLKG DTYGYDKNDF RDPFLFQTED GVYHMLIATR KNGKGHIAEF TSADLKEWES AGTFMTMMWD RF YECPDVF KMGDWWYLIY SEQASFMRKV QYFKGRTLED LKATTANDAG IWPDNREGML DSRAFYAGKT ASDGTNRYIW GWC PTRAGN DNGNVGDVEP EWAGNLVAQR LIQHEDGTLT LGVPDAIDRK YTSAQEVKVM AKDGNMIESG KTYTLGEGAS VIFN RLKVH NKISFTVKTA SNTDRFGISF VRGTDSASWY SIHVNADEGK ANFEKDGDDA KYLFDNKFNI PADNEYRVTI YSDQS VCVT YINDQLSFTN RIYQMQKNPW SLCCYKGEIT VSDVQVSTY UniProtKB: beta-fructofuranosidase |
-Macromolecule #2: DUF4960 domain-containing protein
| Macromolecule | Name: DUF4960 domain-containing protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria)Strain: ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50 |
| Molecular weight | Theoretical: 50.429227 KDa |
| Recombinant expression | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
| Sequence | String: MKSIIKQLYT ILLVTVACLT VTGCSDDFKS GLRLDGDVWV NSIRLDEYAG TVDYQNKAIV VGVPYDYDIT RMVVTEMNLS EGAKASIAI GETIDFSLPV SLTVKNGDVQ MSYTITVKRD EAKILTFKLN DTYVGKVDQL SKTISVVVPL TVDITQLKGT F TVTDGATV ...String: MKSIIKQLYT ILLVTVACLT VTGCSDDFKS GLRLDGDVWV NSIRLDEYAG TVDYQNKAIV VGVPYDYDIT RMVVTEMNLS EGAKASIAI GETIDFSLPV SLTVKNGDVQ MSYTITVKRD EAKILTFKLN DTYVGKVDQL SKTISVVVPL TVDITQLKGT F TVTDGATV TPASGSIQDF TNPVTYTATY RSAVTPYVVT VTQGNVIPTA FVGTASSVSL LTSPEEKAAA QWMMDNVSMS EY ISFKDVV DGKVDLGKYT AIWWHFHADN GDNPPLPDDA KAAAEKFKVY YQNGGNLLLT RYATFYIANL GIAKDERVPN NSW GGNEDS PEITSAPWSF LITGSESHPL FQDLRWKDGD KSTVYTCDAG YAITNSTAQW HIGTDWGGYD DLNAWRNLTG GIDL AHGGD GAVVIAEFEP RSNSGRTLCI GSGCYDWYGK GVDASADYYH YNVEQMTLNA INYLCK UniProtKB: DUF4960 domain-containing protein |
-Macromolecule #3: SusD homolog
| Macromolecule | Name: SusD homolog / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) / Strain: ATCC 29148 / DSM 2079 / NCTC 10582 / E50 / VPI-5482 Bacteroides thetaiotaomicron VPI-5482 (bacteria) / Strain: ATCC 29148 / DSM 2079 / NCTC 10582 / E50 / VPI-5482 |
| Molecular weight | Theoretical: 66.142625 KDa |
| Recombinant expression | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
| Sequence | String: MKKIIYIATI GITLLTTSCD DFLDRQVPQG IVTGDQIASP EYVDNLVISA YAIWATGDDI NSSFSLWNYD VRSDDCYKGG SGTEDGGVF NALEISKGIN TTDWNINDIW KRLYQCITRA NTALQSLDQM DEKTYPLKNQ RIAEMRFLRG HAHFMLKQLF K KIVIVNDE ...String: MKKIIYIATI GITLLTTSCD DFLDRQVPQG IVTGDQIASP EYVDNLVISA YAIWATGDDI NSSFSLWNYD VRSDDCYKGG SGTEDGGVF NALEISKGIN TTDWNINDIW KRLYQCITRA NTALQSLDQM DEKTYPLKNQ RIAEMRFLRG HAHFMLKQLF K KIVIVNDE NMEPDAYNEL SNTTYTNDEQ WQKIADDFQF AYDNLPEVQI EKGRPAQAAA AAYLAKTYLY KAYRQDGADN AL TGINEED LKQVVKYTDP LIMAKGGYGL ETDYSMNFLP QYENGAESVW AIQYSINDGT YNGNLNWGMG LTTPQILGCC DFH KPSQNL VNAFKTDSQG KPLFSTYDNE NYEVATDNVD PRLFHTVGMP GFPYKYNEGY IIQKNDDWSR SKGLYGYYVS LKEN VDPDC DCLKKGSYWA SSLNHIVIRY ADVLLMRAEA LIQLNDGRIT DAISLINEVR SRAAGSTMLI FNYKEDYGVN FKVTP YDLK AYAQDEAMKM LKWERRVEFG MESSRFFDLV RWGEAKDVIN AYYVTEASRC SIYKNAGFTE NKNEYLPVPF EQISAS NGN YTQNFGWAAA AHHHHHH UniProtKB: SusD homolog |
-Macromolecule #4: SusC homolog
| Macromolecule | Name: SusC homolog / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) / Strain: ATCC 29148 / DSM 2079 / NCTC 10582 / E50 / VPI-5482 Bacteroides thetaiotaomicron VPI-5482 (bacteria) / Strain: ATCC 29148 / DSM 2079 / NCTC 10582 / E50 / VPI-5482 |
| Molecular weight | Theoretical: 115.483602 KDa |
| Recombinant expression | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
| Sequence | String: MPGIMKNKKL LCSVCFLFAF MSALWGQNIT VKGNVTSKTD GQPIIGASVV ETTATTNGTI TDFDGNFTLS VPVNSTLKIT YIGYKPVTV KAAAIVNVLL EEDTQMVDEV VVTGYTTQRK ADLTGAVSVV KVDEIQKQGE NNPVKALQGR VPGMNITADG N PSGSATVR ...String: MPGIMKNKKL LCSVCFLFAF MSALWGQNIT VKGNVTSKTD GQPIIGASVV ETTATTNGTI TDFDGNFTLS VPVNSTLKIT YIGYKPVTV KAAAIVNVLL EEDTQMVDEV VVTGYTTQRK ADLTGAVSVV KVDEIQKQGE NNPVKALQGR VPGMNITADG N PSGSATVR IRGIGTLNNN DPLYIIDGVP TKAGMHELNG NDIESIQVLK DAASASIYGS RAANGVIIIT TKQGKKGQIK IN FDASVSA SMYQSKMNVL NTEQYGRAMW QAYVNDGENP NGNALGYAYN WGYNADGNPV LYGMTLSKYL DSKNTMPVAD TDW FDEITR TGVIQQYNLS VSNGSEKGSS FFSLGYYKNL GVIKDTDFDR FSARMNSDYK LIDDILTIGQ HFTLNRTSEV QAPG GIIET ALDIPSAIPV YASDGSWGGP VGGWPDRRNP RAVLEYNKDN RYTYWRMFGD AYVNLTPFKG FNLRSTFGLD YANKQ ARYF TYPYQEGTQT NNGKSAVEAK QEHWTKWMWN AIATYQLEVG KHRGDVMIGM ELNREDDSHF SGYKEDFSIL TPDYMW PDA GSGTAQAYGA GEGYSLVSFF GKMNYSYADR YLLSLTLRRD GSSRFGKNHR YATFPSVSLG WRITQENFMK ELTWLDD LK LRASWGQTGN QEISNLARYT IYAPNYGTTD SFGGQSYGTA YDITGSNGGG VLPSGFKRNQ IGNDNIKWET TTQTNVGI D FSLFKQSLYG SLEYYYKKAT DILTEMAGVG VLGEGGSRWI NSGAMKNQGF EFNLGYRNKT AFGLTYDLNG NISTYRNEI LELPETVAAN GKFGGNGVKS VVGHTYGAQV GYIADGIFKS QDEVDNHATQ EGAAVGRIRY RDIDHNGVID ERDQNWIYDP TPSFSYGLN IYLEYKNFDL TMFWQGVQGV DIISDVKKKS DFWSASNVGF LNKGTRLLNA WSPTNPNSDI PALTRSDTNN E QRVSTYFV ENGSFLKLRN IQLGYTVPAV ISKKMRMDRL RFYCSAQNLL TIKSKNFTGE DPENPNFSYP IPVNITFGLN IG F UniProtKB: SusC homolog |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Supplemented with 0.5 mM levan fructo-oligosaccharides DP 8-12 | ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER / Details: 30 mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 38.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)