[English] 日本語
 Yorodumi
Yorodumi- EMDB-1527: The structure of phosphorylase kinase holoenzyme at 9.9 A resolut... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1527 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of phosphorylase kinase holoenzyme at 9.9 A resolution and location of the catalytic subunit and the substrate glycogen phosphorylase | |||||||||
 Map data Map data | 3D map of PhK | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Phosphorylase kinase | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.9 Å | |||||||||
 Authors Authors | Venien-Bryan C / Jonic S / Skamnaki V / Brown N / Bishler N / Oikonomakos NG / Boisset N / Johnson LN | |||||||||
 Citation Citation |  Journal: Structure / Year: 2009 Journal: Structure / Year: 2009Title: The structure of phosphorylase kinase holoenzyme at 9.9 angstroms resolution and location of the catalytic subunit and the substrate glycogen phosphorylase. Authors: Catherine Vénien-Bryan / Slavica Jonic / Vasiliki Skamnaki / Nick Brown / Nicolas Bischler / Nikos G Oikonomakos / Nicolas Boisset / Louise N Johnson /  Abstract: Phosphorylase kinase (PhK) coordinates hormonal and neuronal signals to initiate the breakdown of glycogen. The enzyme catalyzes the phosphorylation of inactive glycogen phosphorylase b (GPb), ...Phosphorylase kinase (PhK) coordinates hormonal and neuronal signals to initiate the breakdown of glycogen. The enzyme catalyzes the phosphorylation of inactive glycogen phosphorylase b (GPb), resulting in the formation of active glycogen phosphorylase a. We present a 9.9 angstroms resolution structure of PhK heterotetramer (alphabetagammadelta)4 determined by cryo-electron microscopy single-particle reconstruction. The enzyme has a butterfly-like shape comprising two lobes with 222 symmetry. This three-dimensional structure has allowed us to dock the catalytic gamma subunit to the PhK holoenzyme at a location that is toward the ends of the lobes. We have also determined the structure of PhK decorated with GPb at 18 angstroms resolution, which shows the location of the substrate near the kinase subunit. The PhK preparation contained a number of smaller particles whose structure at 9.8 angstroms resolution was consistent with a proteolysed activated form of PhK that had lost the alpha subunits and possibly the gamma subunits. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1527.map.gz emd_1527.map.gz | 20.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1527-v30.xml emd-1527-v30.xml emd-1527.xml emd-1527.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| Images |  1527.gif 1527.gif | 76.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1527 http://ftp.pdbj.org/pub/emdb/structures/EMD-1527 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1527 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1527 | HTTPS FTP |
-Validation report
| Summary document |  emd_1527_validation.pdf.gz emd_1527_validation.pdf.gz | 209.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1527_full_validation.pdf.gz emd_1527_full_validation.pdf.gz | 208.7 KB | Display | |
| Data in XML |  emd_1527_validation.xml.gz emd_1527_validation.xml.gz | 5.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1527 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1527 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1527 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1527 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1527.map.gz / Format: CCP4 / Size: 21.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1527.map.gz / Format: CCP4 / Size: 21.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map of PhK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
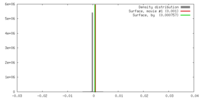
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Phosphorylase kinase holoenzyme purified from rabbit muscle
| Entire | Name: Phosphorylase kinase holoenzyme purified from rabbit muscle |
|---|---|
| Components |
|
-Supramolecule #1000: Phosphorylase kinase holoenzyme purified from rabbit muscle
| Supramolecule | Name: Phosphorylase kinase holoenzyme purified from rabbit muscle type: sample / ID: 1000 Oligomeric state: hexadecamer assembly of four different subunits arranged as an (abgd)4 tetramer Number unique components: 4 |
|---|---|
| Molecular weight | Experimental: 1.3 MDa / Theoretical: 1.3 MDa |
-Macromolecule #1: Phosphorylase kinase
| Macromolecule | Name: Phosphorylase kinase / type: protein_or_peptide / ID: 1 / Name.synonym: PhK / Number of copies: 4 / Oligomeric state: hexadecamer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 1.3 MDa / Theoretical: 1.3 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 8.2 Details: 100mM NaCl, 0.3 mM CaCl2, 5mM MgCl2, 50 mM Hepes, pH 8.2 |
| Grid | Details: 400-mesh copper grid coated with a thin holey-carbon film |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: Manual. 5 microL were applied on a 200 mesh copper grid, coated with a thin holey carbon film. After blotting the excess of solution with Whatman paper, the grid ...Details: Vitrification instrument: Manual. 5 microL were applied on a 200 mesh copper grid, coated with a thin holey carbon film. After blotting the excess of solution with Whatman paper, the grid was rapidly plunged into liquid ethane Method: Single-sided blotting |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010UHR |
|---|---|
| Temperature | Min: 91.15 K / Max: 93.15 K / Average: 93.15 K |
| Details | low dose illumination |
| Date | Apr 13, 2005 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 2.07 µm / Number real images: 98 / Average electron dose: 10 e/Å2 / Bits/pixel: 8 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.5 mm / Nominal defocus max: 4.8 µm / Nominal defocus min: 2.8 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Gatan / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | The particles were selected using an automatic selection program |
|---|---|
| CTF correction | Details: Wiener filtration of volumes from focal series |
| Final reconstruction | Applied symmetry - Point group: D2 (2x2 fold dihedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.9 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER Details: Final map was calculated using five groups of defocus Number images used: 18123 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: Situs |
| Details | Protocol: rigid body |
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT / Target criteria: cross correlation coefficient |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)