[English] 日本語
 Yorodumi
Yorodumi- EMDB-15204: Cryo-EM structure of Listeria monocytogenes 50S ribosomal subunit. -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Listeria monocytogenes 50S ribosomal subunit. | |||||||||
 Map data Map data | Post-processed masked final volume | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / Listeria monocytogenes / 50S | |||||||||
| Function / homology |  Function and homology information Function and homology informationlarge ribosomal subunit / transferase activity / ribosomal large subunit assembly / 5S rRNA binding / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding ...large ribosomal subunit / transferase activity / ribosomal large subunit assembly / 5S rRNA binding / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / RNA binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Listeria monocytogenes EGD-e (bacteria) Listeria monocytogenes EGD-e (bacteria) | |||||||||
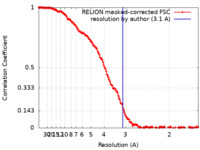
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Koller TO / Crowe-McAuliffe C / Wilson DN | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes. Authors: Timm O Koller / Kathryn J Turnbull / Karolis Vaitkevicius / Caillan Crowe-McAuliffe / Mohammad Roghanian / Ondřej Bulvas / Jose A Nakamoto / Tatsuaki Kurata / Christina Julius / Gemma C ...Authors: Timm O Koller / Kathryn J Turnbull / Karolis Vaitkevicius / Caillan Crowe-McAuliffe / Mohammad Roghanian / Ondřej Bulvas / Jose A Nakamoto / Tatsuaki Kurata / Christina Julius / Gemma C Atkinson / Jörgen Johansson / Vasili Hauryliuk / Daniel N Wilson /      Abstract: HflX is a ubiquitous bacterial GTPase that splits and recycles stressed ribosomes. In addition to HflX, Listeria monocytogenes contains a second HflX homolog, HflXr. Unlike HflX, HflXr confers ...HflX is a ubiquitous bacterial GTPase that splits and recycles stressed ribosomes. In addition to HflX, Listeria monocytogenes contains a second HflX homolog, HflXr. Unlike HflX, HflXr confers resistance to macrolide and lincosamide antibiotics by an experimentally unexplored mechanism. Here, we have determined cryo-EM structures of L. monocytogenes HflXr-50S and HflX-50S complexes as well as L. monocytogenes 70S ribosomes in the presence and absence of the lincosamide lincomycin. While the overall geometry of HflXr on the 50S subunit is similar to that of HflX, a loop within the N-terminal domain of HflXr, which is two amino acids longer than in HflX, reaches deeper into the peptidyltransferase center. Moreover, unlike HflX, the binding of HflXr induces conformational changes within adjacent rRNA nucleotides that would be incompatible with drug binding. These findings suggest that HflXr confers resistance using an allosteric ribosome protection mechanism, rather than by simply splitting and recycling antibiotic-stalled ribosomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15204.map.gz emd_15204.map.gz | 49.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15204-v30.xml emd-15204-v30.xml emd-15204.xml emd-15204.xml | 53.2 KB 53.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15204_fsc.xml emd_15204_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_15204.png emd_15204.png | 80.8 KB | ||
| Filedesc metadata |  emd-15204.cif.gz emd-15204.cif.gz | 10.4 KB | ||
| Others |  emd_15204_additional_1.map.gz emd_15204_additional_1.map.gz emd_15204_half_map_1.map.gz emd_15204_half_map_1.map.gz emd_15204_half_map_2.map.gz emd_15204_half_map_2.map.gz | 224.6 MB 225.4 MB 225.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15204 http://ftp.pdbj.org/pub/emdb/structures/EMD-15204 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15204 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15204 | HTTPS FTP |
-Related structure data
| Related structure data |  8a63MC  8a57C  8a5iC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15204.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15204.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed masked final volume | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
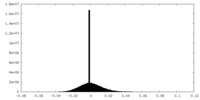
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: final 3D refined volume
| File | emd_15204_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | final 3D refined volume | ||||||||||||
| Projections & Slices |
| ||||||||||||
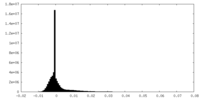
| Density Histograms |
-Half map: Half-map1
| File | emd_15204_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
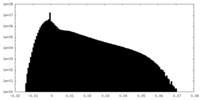
| Density Histograms |
-Half map: Half-map2
| File | emd_15204_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
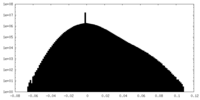
| Density Histograms |
- Sample components
Sample components
+Entire : Listeria monocytogenes 50S ribosomal subunit.
+Supramolecule #1: Listeria monocytogenes 50S ribosomal subunit.
+Macromolecule #1: 50S ribosomal protein L28
+Macromolecule #2: 50S ribosomal protein L29
+Macromolecule #3: 50S ribosomal protein L30
+Macromolecule #4: 50S ribosomal protein L31 type B
+Macromolecule #5: 50S ribosomal protein L32-2
+Macromolecule #6: 50S ribosomal protein L33 1
+Macromolecule #7: 50S ribosomal protein L34
+Macromolecule #8: 50S ribosomal protein L35
+Macromolecule #9: 50S ribosomal protein L36
+Macromolecule #12: 50S ribosomal protein L2
+Macromolecule #13: 50S ribosomal protein L3
+Macromolecule #14: 50S ribosomal protein L4
+Macromolecule #15: 50S ribosomal protein L5
+Macromolecule #16: 50S ribosomal protein L6
+Macromolecule #17: 50S ribosomal protein L13
+Macromolecule #18: 50S ribosomal protein L14
+Macromolecule #19: 50S ribosomal protein L15
+Macromolecule #20: 50S ribosomal protein L16
+Macromolecule #21: 50S ribosomal protein L17
+Macromolecule #22: 50S ribosomal protein L18
+Macromolecule #23: 50S ribosomal protein L19
+Macromolecule #24: 50S ribosomal protein L20
+Macromolecule #25: 50S ribosomal protein L21
+Macromolecule #26: 50S ribosomal protein L22
+Macromolecule #27: 50S ribosomal protein L23
+Macromolecule #28: 50S ribosomal protein L24
+Macromolecule #29: 50S ribosomal protein L27
+Macromolecule #10: 23S ribosomal RNA
+Macromolecule #11: 5S ribosomal RNA
+Macromolecule #30: ZINC ION
+Macromolecule #31: SPERMIDINE
+Macromolecule #32: MAGNESIUM ION
+Macromolecule #33: 1,4-DIAMINOBUTANE
+Macromolecule #34: POTASSIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: 0.05% Nikkol | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 | |||||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||||||||||||||
| Details | 5 OD260/mL |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 30.255 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 165000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)