[English] 日本語
 Yorodumi
Yorodumi- EMDB-15110: Cryo-EM structure of mouse Pannexin 1 purified in Salipro nanopar... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of mouse Pannexin 1 purified in Salipro nanoparticles | |||||||||
 Map data Map data | DeepEMhancer sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane transporter / ATP-release channel / inflammation / immune function / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationElectric Transmission Across Gap Junctions / The NLRP3 inflammasome / ATP transmembrane transporter activity / ATP transport / leak channel activity / positive regulation of interleukin-1 alpha production / bleb / monoatomic anion transmembrane transport / monoatomic anion channel activity / gap junction ...Electric Transmission Across Gap Junctions / The NLRP3 inflammasome / ATP transmembrane transporter activity / ATP transport / leak channel activity / positive regulation of interleukin-1 alpha production / bleb / monoatomic anion transmembrane transport / monoatomic anion channel activity / gap junction / gap junction channel activity / positive regulation of macrophage cytokine production / response to ATP / response to ischemia / positive regulation of interleukin-1 beta production / calcium channel activity / actin filament binding / calcium ion transport / cell-cell signaling / actin binding / protease binding / scaffold protein binding / transmembrane transporter binding / signaling receptor binding / endoplasmic reticulum membrane / structural molecule activity / endoplasmic reticulum / protein-containing complex / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
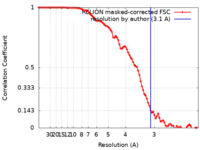
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Drulyte I | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2023 Journal: Sci Rep / Year: 2023Title: Direct cell extraction of membrane proteins for structure-function analysis. Authors: Ieva Drulyte / Aspen Rene Gutgsell / Pilar Lloris-Garcerá / Michael Liss / Stefan Geschwindner / Mazdak Radjainia / Jens Frauenfeld / Robin Löving /    Abstract: Membrane proteins are the largest group of therapeutic targets in a variety of disease areas and yet, they remain particularly difficult to investigate. We have developed a novel one-step approach ...Membrane proteins are the largest group of therapeutic targets in a variety of disease areas and yet, they remain particularly difficult to investigate. We have developed a novel one-step approach for the incorporation of membrane proteins directly from cells into lipid Salipro nanoparticles. Here, with the pannexin1 channel as a case study, we demonstrate the applicability of this method for structure-function analysis using SPR and cryo-EM. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15110.map.gz emd_15110.map.gz | 57 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15110-v30.xml emd-15110-v30.xml emd-15110.xml emd-15110.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15110_fsc.xml emd_15110_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_15110.png emd_15110.png | 124.7 KB | ||
| Filedesc metadata |  emd-15110.cif.gz emd-15110.cif.gz | 6.3 KB | ||
| Others |  emd_15110_additional_1.map.gz emd_15110_additional_1.map.gz emd_15110_half_map_1.map.gz emd_15110_half_map_1.map.gz emd_15110_half_map_2.map.gz emd_15110_half_map_2.map.gz | 48.9 MB 49.4 MB 49.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15110 http://ftp.pdbj.org/pub/emdb/structures/EMD-15110 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15110 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15110 | HTTPS FTP |
-Validation report
| Summary document |  emd_15110_validation.pdf.gz emd_15110_validation.pdf.gz | 669.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15110_full_validation.pdf.gz emd_15110_full_validation.pdf.gz | 668.8 KB | Display | |
| Data in XML |  emd_15110_validation.xml.gz emd_15110_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_15110_validation.cif.gz emd_15110_validation.cif.gz | 21.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15110 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15110 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15110 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15110 | HTTPS FTP |
-Related structure data
| Related structure data |  8a3bMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15110.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15110.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0905 Å | ||||||||||||||||||||||||||||||||||||
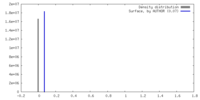
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_15110_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
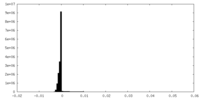
| Density Histograms |
-Half map: Half map 2
| File | emd_15110_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
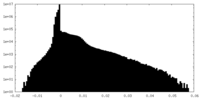
| Density Histograms |
-Half map: Half map 1
| File | emd_15110_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
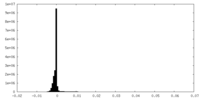
| Density Histograms |
- Sample components
Sample components
-Entire : Salipro-mPANX1
| Entire | Name: Salipro-mPANX1 |
|---|---|
| Components |
|
-Supramolecule #1: Salipro-mPANX1
| Supramolecule | Name: Salipro-mPANX1 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 353 KDa |
-Macromolecule #1: Pannexin-1
| Macromolecule | Name: Pannexin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.475434 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSDYKDDDDK GGGGSMAIAH LATEYVFSDF LLKEPTEPKF KGLRLELAVD KMVTCIAVGL PLLLISLAFA QEISIGTQIS CFSPSSFSW RQAAFVDSYC WAAVQQKSSL QSESGNLPLW LHKFFPYILL LFAILLYLPA LFWRFSAAPH LCSDLKFIME E LDKVYNRA ...String: MSDYKDDDDK GGGGSMAIAH LATEYVFSDF LLKEPTEPKF KGLRLELAVD KMVTCIAVGL PLLLISLAFA QEISIGTQIS CFSPSSFSW RQAAFVDSYC WAAVQQKSSL QSESGNLPLW LHKFFPYILL LFAILLYLPA LFWRFSAAPH LCSDLKFIME E LDKVYNRA IKAAKSARDL DLRDGPGPPG VTENVGQSLW EISESHFKYP IVEQYLKTKK NSSHLIMKYI SCRLVTFVVI LL ACIYLSY YFSLSSLSDE FLCSIKSGVL KNDSTIPDRF QCKLIAVGIF QLLSLINLIV YALLIPVVVY TFFIPFRQKT DIL KVYEIL PTFDVLHFKS EGYNDLSLYN LFLEENISEL KSYKCLKVLE NIKSNGQGID PMLLLTNLGM IKMDIIDGKI PTSL QTKGE DQGSQRVEFK DLDLSSEAAA NNGEKNSRQR LLNPSCLEVL FQ UniProtKB: Pannexin-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 50 mM HEPES at pH 7.5, 150 mM NaCl |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR Details: Grids were glow-discharged using 20 mAmp current for 45 sec and charge set to positive. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: To overcome preferred orientation, 0.5 mM fluorinated Fos-Choline 8 was added to the sample just before the grid freezing. Blot parameter: blot force +20, blot time 10 s, waiting time 30s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Details | Titan Krios G4 was used with fringe-free imaging and aberration-free image shifts. Nominal pixel size for 165kx 0.75 A, calibrated pixel size 0.727 A. |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 7955 / Average exposure time: 4.16 sec. / Average electron dose: 40.24 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8a3b: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)