[English] 日本語
 Yorodumi
Yorodumi- EMDB-15085: RuvAB branch migration motor complexed to the Holliday junction -... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | RuvAB branch migration motor complexed to the Holliday junction - composite map 1 and 2 | |||||||||
 Map data Map data | RuvAB branch migration motor complexed to the Holliday junction - composite map 2 [dataset t2] | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA recombination / DNA repair / branch migration / Holliday junction / helicase / DNA BINDING PROTEIN | |||||||||
| Biological species |  Streptococcus thermophilus (bacteria) Streptococcus thermophilus (bacteria) | |||||||||
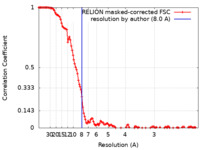
| Method | single particle reconstruction / cryo EM / Resolution: 8.0 Å | |||||||||
 Authors Authors | Wald J / Marlovits TC | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration. Authors: Jiri Wald / Dirk Fahrenkamp / Nikolaus Goessweiner-Mohr / Wolfgang Lugmayr / Luciano Ciccarelli / Oliver Vesper / Thomas C Marlovits /    Abstract: The Holliday junction is a key intermediate formed during DNA recombination across all kingdoms of life. In bacteria, the Holliday junction is processed by two homo-hexameric AAA+ ATPase RuvB motors, ...The Holliday junction is a key intermediate formed during DNA recombination across all kingdoms of life. In bacteria, the Holliday junction is processed by two homo-hexameric AAA+ ATPase RuvB motors, which assemble together with the RuvA-Holliday junction complex to energize the strand-exchange reaction. Despite its importance for chromosome maintenance, the structure and mechanism by which this complex facilitates branch migration are unknown. Here, using time-resolved cryo-electron microscopy, we obtained structures of the ATP-hydrolysing RuvAB complex in seven distinct conformational states, captured during assembly and processing of a Holliday junction. Five structures together resolve the complete nucleotide cycle and reveal the spatiotemporal relationship between ATP hydrolysis, nucleotide exchange and context-specific conformational changes in RuvB. Coordinated motions in a converter formed by DNA-disengaged RuvB subunits stimulate hydrolysis and nucleotide exchange. Immobilization of the converter enables RuvB to convert the ATP-contained energy into a lever motion, which generates the pulling force driving the branch migration. We show that RuvB motors rotate together with the DNA substrate, which, together with a progressing nucleotide cycle, forms the mechanistic basis for DNA recombination by continuous branch migration. Together, our data decipher the molecular principles of homologous recombination by the RuvAB complex, elucidate discrete and sequential transition-state intermediates for chemo-mechanical coupling of hexameric AAA+ motors and provide a blueprint for the design of state-specific compounds targeting AAA+ motors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15085.map.gz emd_15085.map.gz | 4.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15085-v30.xml emd-15085-v30.xml emd-15085.xml emd-15085.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15085_fsc.xml emd_15085_fsc.xml | 12.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_15085.png emd_15085.png | 92.4 KB | ||
| Filedesc metadata |  emd-15085.cif.gz emd-15085.cif.gz | 4.4 KB | ||
| Others |  emd_15085_additional_1.map.gz emd_15085_additional_1.map.gz | 10.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15085 http://ftp.pdbj.org/pub/emdb/structures/EMD-15085 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15085 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15085 | HTTPS FTP |
-Related structure data
| Related structure data |  7pblC  7pbmC  7pbnC  7pboC  7pbpC  7pbqC  7pbrC  7pbsC  7pbtC  7pbuC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15085.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15085.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RuvAB branch migration motor complexed to the Holliday junction - composite map 2 [dataset t2] | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: RuvAB branch migration motor complexed to the Holliday...
| File | emd_15085_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RuvAB branch migration motor complexed to the Holliday junction - composite map 1 [dataset t2] | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RuvAB branch migration motor complexed to the Holliday junction -...
| Entire | Name: RuvAB branch migration motor complexed to the Holliday junction - composite maps [t2] |
|---|---|
| Components |
|
-Supramolecule #1: RuvAB branch migration motor complexed to the Holliday junction -...
| Supramolecule | Name: RuvAB branch migration motor complexed to the Holliday junction - composite maps [t2] type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / Details: RuvAB branch migration complex |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus (bacteria) Streptococcus thermophilus (bacteria) |
| Molecular weight | Theoretical: 650 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.15 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | in-vitro reconstituted freshly before vitrification |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number real images: 30083 / Average exposure time: 5.0 sec. / Average electron dose: 30.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)