+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of murine perforin-2 (Mpeg1) pore in ring form | |||||||||
 Map data Map data | Final map, local resolution filtered | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pore forming protein / MACPF / beta-barrel / immunity / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationdendritic cell antigen processing and presentation / antigen processing and presentation of exogenous peptide antigen / phagolysosome membrane / endolysosome / pore-forming activity / antibacterial innate immune response / wide pore channel activity / antigen processing and presentation of exogenous peptide antigen via MHC class I / phagocytic vesicle / phagocytic vesicle membrane ...dendritic cell antigen processing and presentation / antigen processing and presentation of exogenous peptide antigen / phagolysosome membrane / endolysosome / pore-forming activity / antibacterial innate immune response / wide pore channel activity / antigen processing and presentation of exogenous peptide antigen via MHC class I / phagocytic vesicle / phagocytic vesicle membrane / cytoplasmic vesicle / defense response to Gram-negative bacterium / adaptive immune response / defense response to Gram-positive bacterium / defense response to bacterium Similarity search - Function | |||||||||
| Biological species |  | |||||||||
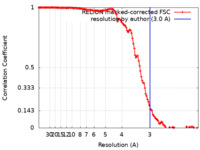
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Yu X / Ni T / Zhang P / Gilbert R | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2022 Journal: EMBO J / Year: 2022Title: Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation. Authors: Xiulian Yu / Tao Ni / George Munson / Peijun Zhang / Robert J C Gilbert /   Abstract: Perforin-2 (PFN2, MPEG1) is a key pore-forming protein in mammalian innate immunity restricting intracellular bacteria proliferation. It forms a membrane-bound pre-pore complex that converts to a ...Perforin-2 (PFN2, MPEG1) is a key pore-forming protein in mammalian innate immunity restricting intracellular bacteria proliferation. It forms a membrane-bound pre-pore complex that converts to a pore-forming structure upon acidification; but its mechanism of conformational transition has been debated. Here we used cryo-electron microscopy, tomography and subtomogram averaging to determine structures of PFN2 in pre-pore and pore conformations in isolation and bound to liposomes. In isolation and upon acidification, the pre-assembled complete pre-pore rings convert to pores in both flat ring and twisted conformations. On membranes, in situ assembled PFN2 pre-pores display various degrees of completeness; whereas PFN2 pores are mainly incomplete arc structures that follow the same subunit packing arrangements as found in isolation. Both assemblies on membranes use their P2 β-hairpin for binding to the lipid membrane surface. Overall, these structural snapshots suggest a molecular mechanism for PFN2 pre-pore to pore transition on a targeted membrane, potentially using the twisted pore as an intermediate or alternative state to the flat conformation, with the capacity to cause bilayer distortion during membrane insertion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15072.map.gz emd_15072.map.gz | 103.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15072-v30.xml emd-15072-v30.xml emd-15072.xml emd-15072.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15072_fsc.xml emd_15072_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15072.png emd_15072.png | 160 KB | ||
| Masks |  emd_15072_msk_1.map emd_15072_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15072.cif.gz emd-15072.cif.gz | 6.7 KB | ||
| Others |  emd_15072_half_map_1.map.gz emd_15072_half_map_1.map.gz emd_15072_half_map_2.map.gz emd_15072_half_map_2.map.gz | 138.8 MB 138.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15072 http://ftp.pdbj.org/pub/emdb/structures/EMD-15072 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15072 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15072 | HTTPS FTP |
-Validation report
| Summary document |  emd_15072_validation.pdf.gz emd_15072_validation.pdf.gz | 838.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15072_full_validation.pdf.gz emd_15072_full_validation.pdf.gz | 838 KB | Display | |
| Data in XML |  emd_15072_validation.xml.gz emd_15072_validation.xml.gz | 20.3 KB | Display | |
| Data in CIF |  emd_15072_validation.cif.gz emd_15072_validation.cif.gz | 26.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15072 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15072 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15072 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15072 | HTTPS FTP |
-Related structure data
| Related structure data |  8a1dMC  8a1sC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15072.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15072.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final map, local resolution filtered | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.047 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15072_msk_1.map emd_15072_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map2 from relion
| File | emd_15072_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map2 from relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map1 from relion
| File | emd_15072_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map1 from relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : murine perforin-2 ectodomain in flat ring pore form
| Entire | Name: murine perforin-2 ectodomain in flat ring pore form |
|---|---|
| Components |
|
-Supramolecule #1: murine perforin-2 ectodomain in flat ring pore form
| Supramolecule | Name: murine perforin-2 ectodomain in flat ring pore form / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Macrophage-expressed gene 1 protein
| Macromolecule | Name: Macrophage-expressed gene 1 protein / type: protein_or_peptide / ID: 1 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 71.359812 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ETGDKPLGET GTTGFQICKN ALKLPVLEVL PGGGWDNLRN VDMGRVMDLT YTNCKTTEDG QYIIPDEVYT IPQKESNLEM NSEVLESWM NYQSTTSLSI NTELALFSRV NGKFSTEFQR MKTLQVKDQA VTTRVQVRNR IYTVKTTPTS ELSLGFTKAL M DICDQLEK ...String: ETGDKPLGET GTTGFQICKN ALKLPVLEVL PGGGWDNLRN VDMGRVMDLT YTNCKTTEDG QYIIPDEVYT IPQKESNLEM NSEVLESWM NYQSTTSLSI NTELALFSRV NGKFSTEFQR MKTLQVKDQA VTTRVQVRNR IYTVKTTPTS ELSLGFTKAL M DICDQLEK NQTKMATYLA ELLILNYGTH VITSVDAGAA LVQEDHVRSS FLLDNQNSQN TVTASAGIAF LNIVNFKVET DY ISQTSLT KDYLSNRTNS RVQSFGGVPF YPGITLETWQ KGITNHLVAI DRAGLPLHFF IKPDKLPGLP GPLVKKLSKT VET AVRHYY TFNTHPGCTN VDSPNFNFQA NMDDDSCDAK VTNFTFGGVY QECTELSGDV LCQNLEQKNL LTGDFSCPPG YSPV HLLSQ THEEGYSRLE CKKKCTLKIF CKTVCEDVFR VAKAEFRAYW CVAAGQVPDN SGLLFGGVFT DKTINPMTNA QSCPA GYIP LNLFESLKVC VSLDYELGFK FSVPFGGFFS CIMGNPLVNS DTAKDVRAPS LKKCPGGFSQ HLAVISDGCQ VSYCVK AGI FTGGSLLPVR LPPYTKPPLM SQVATNTVIV TNSETARSWI KDPQTNQWKL GEPLELRRAM TVIHGDSNGM SGGGTKT ET SQVAPA UniProtKB: Macrophage-expressed gene 1 protein |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 16 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #3: CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE
| Macromolecule | Name: CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE / type: ligand / ID: 3 / Number of copies: 16 / Formula: MA4 |
|---|---|
| Molecular weight | Theoretical: 508.6 Da |
| Chemical component information | 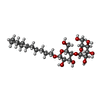 ChemComp-MA4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 3.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-8a1d: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)