+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BcsHD cis-filaments: four 'beads-on-a-string' | |||||||||
 Map data Map data | BcsHD 'beads-on-a-string' filaments : Unsharpened locally refined map of a BcsH-bound tetramer of BcsD octamers | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Komagataeibacter hansenii ATCC 23769 (bacteria) Komagataeibacter hansenii ATCC 23769 (bacteria) | |||||||||
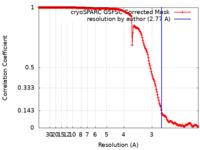
| Method | single particle reconstruction / cryo EM / Resolution: 2.77 Å | |||||||||
 Authors Authors | Krasteva PV / Abidi W / Decossas M | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Bacterial crystalline cellulose secretion via a supramolecular BcsHD scaffold. Authors: Wiem Abidi / Marion Decossas / Lucía Torres-Sánchez / Lucie Puygrenier / Sylvie Létoffé / Jean-Marc Ghigo / Petya V Krasteva /  Abstract: Cellulose, the most abundant biopolymer on Earth, is not only the predominant constituent of plants but also a key extracellular polysaccharide in the biofilms of many bacterial species. Depending on ...Cellulose, the most abundant biopolymer on Earth, is not only the predominant constituent of plants but also a key extracellular polysaccharide in the biofilms of many bacterial species. Depending on the producers, chemical modifications, and three-dimensional assemblies, bacterial cellulose (BC) can present diverse degrees of crystallinity. Highly ordered, or crystalline, cellulose presents great economical relevance due to its ever-growing number of biotechnological applications. Even if some acetic acid bacteria have long been identified as BC superproducers, the molecular mechanisms determining the secretion of crystalline versus amorphous cellulose remain largely unknown. Here, we present structural and mechanistic insights into the role of the accessory subunits BcsH (CcpAx) and BcsD (CesD) that determine crystalline BC secretion in the lineage. We show that oligomeric BcsH drives the assembly of BcsD into a supramolecular cytoskeletal scaffold that likely stabilizes the cellulose-extruding synthase nanoarrays through an unexpected inside-out mechanism for secretion system assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15040.map.gz emd_15040.map.gz | 275.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15040-v30.xml emd-15040-v30.xml emd-15040.xml emd-15040.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15040_fsc.xml emd_15040_fsc.xml | 17.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_15040.png emd_15040.png | 48.9 KB | ||
| Masks |  emd_15040_msk_1.map emd_15040_msk_1.map | 548.9 MB |  Mask map Mask map | |
| Others |  emd_15040_additional_1.map.gz emd_15040_additional_1.map.gz emd_15040_half_map_1.map.gz emd_15040_half_map_1.map.gz emd_15040_half_map_2.map.gz emd_15040_half_map_2.map.gz | 518.8 MB 509.1 MB 509.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15040 http://ftp.pdbj.org/pub/emdb/structures/EMD-15040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15040 | HTTPS FTP |
-Validation report
| Summary document |  emd_15040_validation.pdf.gz emd_15040_validation.pdf.gz | 619.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15040_full_validation.pdf.gz emd_15040_full_validation.pdf.gz | 619.1 KB | Display | |
| Data in XML |  emd_15040_validation.xml.gz emd_15040_validation.xml.gz | 26.9 KB | Display | |
| Data in CIF |  emd_15040_validation.cif.gz emd_15040_validation.cif.gz | 35.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15040 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15040 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15040 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15040 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15040.map.gz / Format: CCP4 / Size: 548.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15040.map.gz / Format: CCP4 / Size: 548.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BcsHD 'beads-on-a-string' filaments : Unsharpened locally refined map of a BcsH-bound tetramer of BcsD octamers | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05676 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15040_msk_1.map emd_15040_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: BcsHD 'beads-on-a-string' filaments :Autosharpened locally refined map of...
| File | emd_15040_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BcsHD 'beads-on-a-string' filaments :Autosharpened locally refined map of a BcsH-bound tetramer of BcsD octamers | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: BcsHD 'beads-on-a-string' filaments : half-map B
| File | emd_15040_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BcsHD 'beads-on-a-string' filaments : half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: BcsHD 'beads-on-a-string' filaments : half-map A
| File | emd_15040_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BcsHD 'beads-on-a-string' filaments : half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BcsHD 'beads-on-a-string' cis-filaments
| Entire | Name: BcsHD 'beads-on-a-string' cis-filaments |
|---|---|
| Components |
|
-Supramolecule #1: BcsHD 'beads-on-a-string' cis-filaments
| Supramolecule | Name: BcsHD 'beads-on-a-string' cis-filaments / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all Details: BcsHD filaments purified upon coexpression of BcsD (no tag) and BcsH C-terminal region carrying an HRV3c-cleavable N-terminal hexahistidine tag. |
|---|---|
| Source (natural) | Organism:  Komagataeibacter hansenii ATCC 23769 (bacteria) / Strain: ATCC 23769 / Location in cell: cytoskeleton Komagataeibacter hansenii ATCC 23769 (bacteria) / Strain: ATCC 23769 / Location in cell: cytoskeleton |
-Macromolecule #1: BcsHD 'beads-on-a-string' cis-filaments
| Macromolecule | Name: BcsHD 'beads-on-a-string' cis-filaments / type: other / ID: 1 Details: Multiple copies of BcsD and BcsH (protein in sample is tag-free with sequence: PMGSTKTDTNSSQASRPGSPVASPDGSPTMAEVFMTLGGRATELLSPRPSLREALLRRRENEEES) in BcsH-driven cis-filaments Classification: other |
|---|---|
| Source (natural) | Organism:  Komagataeibacter hansenii ATCC 23769 (bacteria) / Strain: ATCC 23769 Komagataeibacter hansenii ATCC 23769 (bacteria) / Strain: ATCC 23769 |
| Sequence | String: BcsD MGSTK TDTNSSQASR PGSPVASPDG SPTMAEVFMT LGGRATELLS PRPSLREALL RRRENEEES BcsH MSYYH HHHHHDYDIP TTLEVLFQGP MGSTKTDTNS SQASRPGSPV ASPDGSPTMA EVFMTLGGRA TELLSPRPSL REALLRRREN EEES |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM HEPES pH 8.0, 100 mM NaCl |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 20 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 49.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.75 µm / Nominal defocus min: 0.48 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)