[English] 日本語
 Yorodumi
Yorodumi- EMDB-14996: Structure of SNAPc:TBP-TFIIA-TFIIB sub-complex bound to U5 snRNA ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of SNAPc:TBP-TFIIA-TFIIB sub-complex bound to U5 snRNA promoter | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | RNA polymerase II / Pol II / PIC / SNAPc / snRNA / U5 promoter / TRANSCRIPTION | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsnRNA-activating protein complex / snRNA transcription / snRNA transcription by RNA polymerase III / bent DNA binding / positive regulation of core promoter binding / RNA polymerase II core complex assembly / RNA polymerase transcription factor SL1 complex / RNA polymerase III type 3 promoter sequence-specific DNA binding / meiotic sister chromatid cohesion / snRNA transcription by RNA polymerase II ...snRNA-activating protein complex / snRNA transcription / snRNA transcription by RNA polymerase III / bent DNA binding / positive regulation of core promoter binding / RNA polymerase II core complex assembly / RNA polymerase transcription factor SL1 complex / RNA polymerase III type 3 promoter sequence-specific DNA binding / meiotic sister chromatid cohesion / snRNA transcription by RNA polymerase II / RNA polymerase III general transcription initiation factor activity / transcriptional start site selection at RNA polymerase II promoter / RNA polymerase I core promoter sequence-specific DNA binding / RNA Polymerase III Transcription Initiation From Type 1 Promoter / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Polymerase III Transcription Initiation From Type 3 Promoter / transcription factor TFIIA complex / female germ cell nucleus / RNA Polymerase III Abortive And Retractive Initiation / male pronucleus / female pronucleus / germinal vesicle / RNA polymerase II general transcription initiation factor binding / nuclear thyroid hormone receptor binding / transcription preinitiation complex / RNA Polymerase I Transcription Termination / protein acetylation / RNA polymerase II general transcription initiation factor activity / transcription factor TFIID complex / cell division site / acetyltransferase activity / HIV Transcription Initiation / RNA Polymerase II HIV Promoter Escape / Transcription of the HIV genome / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / transcription initiation at RNA polymerase III promoter / aryl hydrocarbon receptor binding / RNA polymerase II complex binding / viral transcription / TFIIB-class transcription factor binding / RNA Polymerase I Transcription Initiation / transcription by RNA polymerase III / RNA polymerase II transcribes snRNA genes / positive regulation of transcription initiation by RNA polymerase II / histone acetyltransferase activity / RNA polymerase II core promoter sequence-specific DNA binding / core promoter sequence-specific DNA binding / histone acetyltransferase / RNA polymerase II preinitiation complex assembly / spindle assembly / RNA Polymerase II Pre-transcription Events / TBP-class protein binding / SIRT1 negatively regulates rRNA expression / male germ cell nucleus / transcription initiation at RNA polymerase II promoter / promoter-specific chromatin binding / RNA Polymerase I Promoter Escape / DNA-templated transcription initiation / mRNA transcription by RNA polymerase II / euchromatin / protein-DNA complex / NoRC negatively regulates rRNA expression / B-WICH complex positively regulates rRNA expression / kinetochore / RNA polymerase II transcription regulator complex / chromosome / spermatogenesis / Regulation of TP53 Activity through Phosphorylation / Estrogen-dependent gene expression / sequence-specific DNA binding / DNA-binding transcription factor binding / RNA polymerase II-specific DNA-binding transcription factor binding / transcription by RNA polymerase II / transcription cis-regulatory region binding / nuclear body / RNA polymerase II cis-regulatory region sequence-specific DNA binding / protein heterodimerization activity / chromatin / nucleolus / enzyme binding / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / protein-containing complex / DNA binding / zinc ion binding / nucleoplasm / nucleus / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
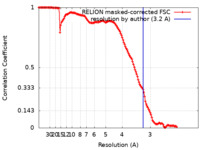
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||
 Authors Authors | Rengachari S / Schilbach S / Kaliyappan T / Gouge J / Zumer K / Schwarz J / Urlaub H / Dienemann C / Vannini A / Cramer P | |||||||||||||||
| Funding support |  Germany, Germany,  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II. Authors: Srinivasan Rengachari / Sandra Schilbach / Thangavelu Kaliyappan / Jerome Gouge / Kristina Zumer / Juliane Schwarz / Henning Urlaub / Christian Dienemann / Alessandro Vannini / Patrick Cramer /    Abstract: RNA polymerase II (Pol II) carries out transcription of both protein-coding and non-coding genes. Whereas Pol II initiation at protein-coding genes has been studied in detail, Pol II initiation at ...RNA polymerase II (Pol II) carries out transcription of both protein-coding and non-coding genes. Whereas Pol II initiation at protein-coding genes has been studied in detail, Pol II initiation at non-coding genes, such as small nuclear RNA (snRNA) genes, is less well understood at the structural level. Here, we study Pol II initiation at snRNA gene promoters and show that the snRNA-activating protein complex (SNAPc) enables DNA opening and transcription initiation independent of TFIIE and TFIIH in vitro. We then resolve cryo-EM structures of the SNAPc-containing Pol IIpre-initiation complex (PIC) assembled on U1 and U5 snRNA promoters. The core of SNAPc binds two turns of DNA and recognizes the snRNA promoter-specific proximal sequence element (PSE), located upstream of the TATA box-binding protein TBP. Two extensions of SNAPc, called wing-1 and wing-2, bind TFIIA and TFIIB, respectively, explaining how SNAPc directs Pol II to snRNA promoters. Comparison of structures of closed and open promoter complexes elucidates TFIIH-independent DNA opening. These results provide the structural basis of Pol II initiation at non-coding RNA gene promoters. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14996.map.gz emd_14996.map.gz | 203.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14996-v30.xml emd-14996-v30.xml emd-14996.xml emd-14996.xml | 31.8 KB 31.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14996_fsc.xml emd_14996_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14996.png emd_14996.png | 161.1 KB | ||
| Masks |  emd_14996_msk_1.map emd_14996_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14996.cif.gz emd-14996.cif.gz | 8.5 KB | ||
| Others |  emd_14996_additional_1.map.gz emd_14996_additional_1.map.gz emd_14996_half_map_1.map.gz emd_14996_half_map_1.map.gz emd_14996_half_map_2.map.gz emd_14996_half_map_2.map.gz | 142.4 MB 195.2 MB 195.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14996 http://ftp.pdbj.org/pub/emdb/structures/EMD-14996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14996 | HTTPS FTP |
-Validation report
| Summary document |  emd_14996_validation.pdf.gz emd_14996_validation.pdf.gz | 862 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14996_full_validation.pdf.gz emd_14996_full_validation.pdf.gz | 861.5 KB | Display | |
| Data in XML |  emd_14996_validation.xml.gz emd_14996_validation.xml.gz | 21.6 KB | Display | |
| Data in CIF |  emd_14996_validation.cif.gz emd_14996_validation.cif.gz | 28.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14996 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14996 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14996 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14996 | HTTPS FTP |
-Related structure data
| Related structure data |  7zwcMC  7zwdC  7zx7C  7zx8C  7zxeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14996.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14996.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14996_msk_1.map emd_14996_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_14996_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14996_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_14996_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : General transcription factors and SNAPc bound to U5 promoter
+Supramolecule #1: General transcription factors and SNAPc bound to U5 promoter
+Supramolecule #2: SNAPc
+Supramolecule #3: TATA-box-binding protein and transcription factors
+Supramolecule #4: Promoter
+Macromolecule #1: Transcription initiation factor IIB
+Macromolecule #2: TATA-box-binding protein
+Macromolecule #3: Transcription initiation factor IIA subunit 1
+Macromolecule #4: Transcription initiation factor IIA subunit 2
+Macromolecule #5: snRNA-activating protein complex subunit 1
+Macromolecule #6: snRNA-activating protein complex subunit 3
+Macromolecule #7: snRNA-activating protein complex subunit 4
+Macromolecule #8: snRNA-activating protein complex subunit 5
+Macromolecule #9: Non-template strand
+Macromolecule #10: Template strand
+Macromolecule #11: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 51.93 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)



















































 Trichoplusia ni (cabbage looper)
Trichoplusia ni (cabbage looper)