+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | HOPS tethering complex from yeast, composite map | ||||||||||||
 Map data Map data | Tethering complex HOPS, composite map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HOPS / tethering complex / lysosome / membrane fusion / Rab GTPase / cryo-EM / CYTOSOLIC PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationhistone catabolic process / organelle fusion / CORVET complex / HOPS complex / endosomal vesicle fusion / vesicle tethering / regulation of vacuole fusion, non-autophagic / vacuole-mitochondrion membrane contact site / vacuolar protein processing / Golgi to vacuole transport ...histone catabolic process / organelle fusion / CORVET complex / HOPS complex / endosomal vesicle fusion / vesicle tethering / regulation of vacuole fusion, non-autophagic / vacuole-mitochondrion membrane contact site / vacuolar protein processing / Golgi to vacuole transport / regulation of SNARE complex assembly / vesicle fusion with vacuole / vacuole fusion, non-autophagic / cytoplasm to vacuole targeting by the Cvt pathway / Golgi to endosome transport / vesicle docking / vacuole organization / protein targeting to vacuole / late endosome to vacuole transport / endosome organization / piecemeal microautophagy of the nucleus / fungal-type vacuole / fungal-type vacuole membrane / vesicle docking involved in exocytosis / endosomal transport / vesicle-mediated transport / positive regulation of TORC1 signaling / guanyl-nucleotide exchange factor activity / cellular response to starvation / macroautophagy / intracellular protein transport / RING-type E3 ubiquitin transferase / autophagy / small GTPase binding / endocytosis / ubiquitin protein ligase activity / late endosome / protein transport / actin binding / early endosome membrane / protein-macromolecule adaptor activity / endosome / zinc ion binding / ATP binding / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | ||||||||||||
 Authors Authors | Shvarev D / Schoppe J / Koenig C / Perz A / Fuellbrunn N / Kiontke S / Langemeyer L / Januliene D / Schnelle K / Kuemmel D ...Shvarev D / Schoppe J / Koenig C / Perz A / Fuellbrunn N / Kiontke S / Langemeyer L / Januliene D / Schnelle K / Kuemmel D / Froehlich F / Moeller A / Ungermann C | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Structure of the HOPS tethering complex, a lysosomal membrane fusion machinery. Authors: Dmitry Shvarev / Jannis Schoppe / Caroline König / Angela Perz / Nadia Füllbrunn / Stephan Kiontke / Lars Langemeyer / Dovile Januliene / Kilian Schnelle / Daniel Kümmel / Florian ...Authors: Dmitry Shvarev / Jannis Schoppe / Caroline König / Angela Perz / Nadia Füllbrunn / Stephan Kiontke / Lars Langemeyer / Dovile Januliene / Kilian Schnelle / Daniel Kümmel / Florian Fröhlich / Arne Moeller / Christian Ungermann /  Abstract: Lysosomes are essential for cellular recycling, nutrient signaling, autophagy, and pathogenic bacteria and viruses invasion. Lysosomal fusion is fundamental to cell survival and requires HOPS, a ...Lysosomes are essential for cellular recycling, nutrient signaling, autophagy, and pathogenic bacteria and viruses invasion. Lysosomal fusion is fundamental to cell survival and requires HOPS, a conserved heterohexameric tethering complex. On the membranes to be fused, HOPS binds small membrane-associated GTPases and assembles SNAREs for fusion, but how the complex fulfills its function remained speculative. Here, we used cryo-electron microscopy to reveal the structure of HOPS. Unlike previously reported, significant flexibility of HOPS is confined to its extremities, where GTPase binding occurs. The SNARE-binding module is firmly attached to the core, therefore, ideally positioned between the membranes to catalyze fusion. Our data suggest a model for how HOPS fulfills its dual functionality of tethering and fusion and indicate why it is an essential part of the membrane fusion machinery. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14964.map.gz emd_14964.map.gz | 1.1 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14964-v30.xml emd-14964-v30.xml emd-14964.xml emd-14964.xml | 30.6 KB 30.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14964.png emd_14964.png | 75.6 KB | ||
| Filedesc metadata |  emd-14964.cif.gz emd-14964.cif.gz | 10.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14964 http://ftp.pdbj.org/pub/emdb/structures/EMD-14964 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14964 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14964 | HTTPS FTP |
-Validation report
| Summary document |  emd_14964_validation.pdf.gz emd_14964_validation.pdf.gz | 518.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14964_full_validation.pdf.gz emd_14964_full_validation.pdf.gz | 518.2 KB | Display | |
| Data in XML |  emd_14964_validation.xml.gz emd_14964_validation.xml.gz | 9 KB | Display | |
| Data in CIF |  emd_14964_validation.cif.gz emd_14964_validation.cif.gz | 10.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14964 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14964 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14964 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14964 | HTTPS FTP |
-Related structure data
| Related structure data |  7zu0MC  7ztyC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14964.map.gz / Format: CCP4 / Size: 1.1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14964.map.gz / Format: CCP4 / Size: 1.1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tethering complex HOPS, composite map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.924 Å | ||||||||||||||||||||||||||||||||||||
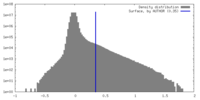
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Tethering complex HOPS
| Entire | Name: Tethering complex HOPS |
|---|---|
| Components |
|
-Supramolecule #1: Tethering complex HOPS
| Supramolecule | Name: Tethering complex HOPS / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: E3 ubiquitin-protein ligase PEP5
| Macromolecule | Name: E3 ubiquitin-protein ligase PEP5 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 117.617219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLSSWRQFQ LFENIPIRDP NFGGDSLLYS DPTLCAATIV DPQTLIIAVN SNIIKVVKLN QSQVIHEFQS FPHDFQITFL KVINGEFLV ALAESIGKPS LIRVYKLEKL PNREQLYHSQ VELKNGNNTY PISVVSISND LSCIVVGFIN GKIILIRGDI S RDRGSQQR ...String: MSLSSWRQFQ LFENIPIRDP NFGGDSLLYS DPTLCAATIV DPQTLIIAVN SNIIKVVKLN QSQVIHEFQS FPHDFQITFL KVINGEFLV ALAESIGKPS LIRVYKLEKL PNREQLYHSQ VELKNGNNTY PISVVSISND LSCIVVGFIN GKIILIRGDI S RDRGSQQR IIYEDPSKEP ITALFLNNDA TACFAATTSR ILLFNTTGRN RGRPSLVLNS KNGLDLNCGS FNPATNEFIC CL SNFIEFF SSSGKKHQFA FDLSLRKRIF CVDKDHILIV TEETGVPTTS ISVNELSPTI INRIFIIDAK NKIISLNFVV SSA IIDIFS TSQSGKNITY LLTSEGVMHR ITPKSLENQI NIIIQKELYP FALQLAKQHS LSPLDVQEIH KKYGDYLFKK GLRK EATDQ YIQCLDVVET SEIISKFGVK EVPDPESMRN LADYLWSLIK NSISQRDHVT LLLIVLIKLK DVEGIDTFIQ HFDRK GIWN EGVVMDDMDD VTFFYSDNDF FDLDLILELM KESDFKRLSY RLAKKYSKDS LIIVDILLNL LHNPVKAIKY IKSLPI DET LRCLVTYSKK LLEESPNETN ALLIEVFTGK FKPSTFEVDL DRRDTTGDFS ENIRTVFYSY KTFFNYMNSN GTSDAMS ES SEASHEHEEP TYHPPKPSIV FSSFVTKPFE FVVFLEACLA CYQQYEGFDE DRQVILTTLY DLYLNLAQND VPERIDDW R SRATGVLRES NKLVYSAASN NTSKRVDNSI MLLISHMDQS SASAKDKTKI DIASFANDNP EMDLLSTFRA MTLNEEPST CLKFLEKYGT EEPKLLQVAL SYFVSNKLIF KEMGGNEVLK EKVLRPIIEG ERMPLLDIIK ALSRTNVAHF GLIQDIIIDH VKTEDTEIK RNEKLIESYD KELKEKNKKL KNTINSDQPL HVPLKNQTCF MCRLTLDIPV VFFKCGHIYH QHCLNEEEDT L ESERKLFK CPKCLVDLET SNKLFEAQHE VVEKNDLLNF ALNSEEGSRD RFKVITEFLG RGAISYSDIT I UniProtKB: E3 ubiquitin-protein ligase PEP5 |
-Macromolecule #2: Vacuolar protein sorting-associated protein 16
| Macromolecule | Name: Vacuolar protein sorting-associated protein 16 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.857 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKNPSFDWER LKDVFYRSRA IGELKWPTQY EEFKCALSLT VIAVEIQDFI QVYNYFGQLL GKINLQRIHE DIIKFEFDKD EKLILVTKS SIKIVKGWSP LTIESVPLQD PTIDTIWDYH NGIMLLAKSR DIYKLNGNEW ELLYENKDKK YNLLTKNHWS C NDDSIILL ...String: MKNPSFDWER LKDVFYRSRA IGELKWPTQY EEFKCALSLT VIAVEIQDFI QVYNYFGQLL GKINLQRIHE DIIKFEFDKD EKLILVTKS SIKIVKGWSP LTIESVPLQD PTIDTIWDYH NGIMLLAKSR DIYKLNGNEW ELLYENKDKK YNLLTKNHWS C NDDSIILL DVDHVYQVST SNGALLKLIT DSSWHKVTIS SRGFICLYNM KDNKLQIFRD PARILMEHNL DSTPDDICWC GN DTVACSF EDEIKLYGPD GLYVTFWYPF TVTNLRAEVD GLKVITTEKI YFLSRVQPQT SNIFRIGSTE PGAMLVDSFS LLE DHAPKA IEILKNFVLE KGVLDCIAAA IDEFEPKLQK MLLNAASYGK ASLQYKSFDA SIFVNACNTI KLLNCFRSFG IFLT VEEYR CISLKGVIDR LLKYHRYYEC IQICKLANER FLLGYVFTEW AKDKIKGSPD MEDDELLDKI KSRLSVIDMT DTLQM VAVA KVAYLEGRFQ LSRNLALLEK NEEARIEQLY NLDDDSIALK ECIKVQNYSL TISLLIALSK KLTNSQLTKL LIIDMF NNP LYLYYMRMDK AYLYDFYRQT DRFIDLAHVL LQQGKEQQSL HSFLPQIKDL YSQVQNSEVV NNTIEQLQRQ EKLWIYQ ES LGKRFAISFT NMTLDQTLSK LIETGQDKQV KEIVKKFKIS EKKLYHLKCK TLVEAKKFDE LLQFAQSRKS PIGYMPFY T YLKSRGHMDK ASPYVNMIPG LSYQEKKKLY VECRGFRDAI QLAGKEKDIP GLKEIYNIIP PNEPELKALA NETMSRI UniProtKB: Vacuolar protein sorting-associated protein 16 |
-Macromolecule #3: Vacuolar membrane protein PEP3
| Macromolecule | Name: Vacuolar membrane protein PEP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 107.531047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIKTRIEEVQ LQFLTGNTEL THLKVSNDQL IVTTQRTIYR INLQDPAIVN HFDCPLSKEL ETIMNVHVSP MGSVILIRTN FGRYMLLKD GEFTQLNKIK NLDLSSLHWI NETTFLMGIK KTPKLYRVEL TGKDITTKLW YENKKLSGGI DGIAYWEGSL L LTIKDNIL ...String: MIKTRIEEVQ LQFLTGNTEL THLKVSNDQL IVTTQRTIYR INLQDPAIVN HFDCPLSKEL ETIMNVHVSP MGSVILIRTN FGRYMLLKD GEFTQLNKIK NLDLSSLHWI NETTFLMGIK KTPKLYRVEL TGKDITTKLW YENKKLSGGI DGIAYWEGSL L LTIKDNIL YWRDVTNMKF PLVLPDESEQ FERLKHHAIK KFDSYNGLFA WVTSNGIVFG DLKEKQMEKD PASNNFGKFL SS SKVLLNF ELPDYQNDKD HLIKDIVLTA FHILLLRKNT VTMVSQLNND VVFHETIPRH QLTGSNTDSN EKFLGLVRDS VKE TFWCFS NINVFEIIIE NEPNSVWNLL VRDNKFDKAL SLKGLTVREI ESVKLSKAMY LFHTAKDFHS AAQTLGSMKD LSHF GEIAL NFLQIKDYND LNVILIKQLD NVPWKSTQVV LSSWIIWNFM KQLNDIELKI NTTKPASTDE DNLLNWNLNL KEKSN ELTK FLESHLEKLD NETVYQIMSK QNRQNELLIF ASLINDMKFL LSFWIDQGNW YESLKILLTI NNHDLVYKYS LILLLN SPE ATVSTWMKIK DLDPNKLIPT ILKFFTNWQN NSKLITNISE YPENYSLTYL KWCVREVPKM CNPIVYNSIL YMMITDP RN DMILENDIIK FMKSNENKYD LNFQLRLSLK FKKTKTSIFL LTRLNLFEDA IDLALKNNLI DDCKVIVNDE ILIEDYKL R KRLWLKIAKH LLLSMKDIDI KQLIRTILND SNEILTIKDL LPFFNEYTTI ANLKEELIKF LENHNMKMNE ISEDIINSK NLKVEINTEI SKFNEIYRIL EPGKSCDECG KFLQIKKFIV FPCGHCFHWN CIIRVILNSN DYNLRQKTEN FLKAKSKHNL NDLENIIVE KCGLCSDINI NKIDQPISID ETELAKWNE UniProtKB: Vacuolar membrane protein PEP3 |
-Macromolecule #4: Vacuolar protein sorting-associated protein 33
| Macromolecule | Name: Vacuolar protein sorting-associated protein 33 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 79.354977 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNRFWNTKKF SLTNADGLCA TLNEISQNDE VLVVQPSVLP VLNSLLTFQD LTQSTPVRKI TLLDDQLSDD LPSALGSVPQ MDLIFLIDV RTSLRLPPQL LDAAQKHNLS SLHIIYCRWK PSFQNTLEDT EQWQKDGFDL NSKKTHFPNV IESQLKELSN E YTLYPWDL ...String: MNRFWNTKKF SLTNADGLCA TLNEISQNDE VLVVQPSVLP VLNSLLTFQD LTQSTPVRKI TLLDDQLSDD LPSALGSVPQ MDLIFLIDV RTSLRLPPQL LDAAQKHNLS SLHIIYCRWK PSFQNTLEDT EQWQKDGFDL NSKKTHFPNV IESQLKELSN E YTLYPWDL LPFPQIDENV LLTHSLYNME NVNMYYPNLR SLQSATESIL VDDMVNSLQS LIFETNSIIT NVVSIGNLSK RC SHLLKKR IDEHQTENDL FIKGTLYGER TNCGLEMDLI ILERNTDPIT PLLTQLTYAG ILDDLYEFNS GIKIKEKDMN FNY KEDKIW NDLKFLNFGS IGPQLNKLAK ELQTQYDTRH KAESVHEIKE FVDSLGSLQQ RQAFLKNHTT LSSDVLKVVE TEEY GSFNK ILELELEILM GNTLNNDIED IILELQYQYE VDQKKILRLI CLLSLCKNSL REKDYEYLRT FMIDSWGIEK CFQLE SLAE LGFFTSKTGK TDLHITTSKS TRLQKEYRYI SQWFNTVPIE DEHAADKITN ENDDFSEATF AYSGVVPLTM RLVQML YDR SILFHNYSSQ QPFILSREPR VSQTEDLIEQ LYGDSHAIEE SIWVPGTITK KINASIKSNN RRSIDGSNGT FHAAEDI AL VVFLGGVTMG EIAIMKHLQK ILGKKGINKR FIIIADGLIN GTRIMNSIS UniProtKB: Vacuolar protein sorting-associated protein 33 |
-Macromolecule #5: Vacuolar morphogenesis protein 6
| Macromolecule | Name: Vacuolar morphogenesis protein 6 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 123.049414 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLRAQKLHSL KSSDITAILP TEQSQKLVLA KKNGDVEVYS RDGNTLKLFQ VYPDLLQNAK NDPLPPVIEN FYFANELSTI FAQCKETLI LLSTTNLHEY DRIIDRRGIN HCWLFERSHK NKEEKNTYLI YSTINTAKMR VLIWEGRTYK NMMEASLSYR K ETIRSIYP ...String: MLRAQKLHSL KSSDITAILP TEQSQKLVLA KKNGDVEVYS RDGNTLKLFQ VYPDLLQNAK NDPLPPVIEN FYFANELSTI FAQCKETLI LLSTTNLHEY DRIIDRRGIN HCWLFERSHK NKEEKNTYLI YSTINTAKMR VLIWEGRTYK NMMEASLSYR K ETIRSIYP GETGITLATD LGIYHWPYNK PSLIRIEKTV KNKFPKDMIS ALTELKEQAE KVIEKKPKKN SHFDAQSFSS MD RMSRKSS MSSLWYRTIR NERGNKIRYT FELDGNDATP MIIDGATKKI FKVELMHNNE EPFLIATDHA TFSESNSEFD HMQ YLSSNL LMLYNSSTIK FVDYENGFTF LQQKIPEGIK WVKNLSGTYF LVWTSNDEVQ LFSYHVDDGS EDDDQESICG DIND PDFYQ LWRKVLFYKF FIDSPHSKEL CVSDNPEESL DICAMKLRDL TVMWCLRIFD KFQNYMVQLE RSRNSRMIRS KCEEM IIKS IFDLFIKFWA PPQLVILKVF PSAISSLVLE ITGQEHHCLL KEAEEVKETY DIPPHLLNRW CLPYLTDTRR HLQNLL SKE NDDESRITWC YRDREIKQSF DFFLISNHDD VDLNTMLTLI DTVLFKCYLY YNPPMVGPFI RVENHCDSHV IVTELKI RH MFKDLIDFYY KRGNHEEALK FLTDLVDELE NDNTDQKQRQ KIDHGVKILV IYYLKKLSNP QLDVIFTYTD WLLNRHND S IKEILSSIFF YDSQACSSRD HLKVYGYIKK FDKLLAIQYL EFAISTFRLE GNKLHTVLIK LYLENLDIPS TRIKLKSLL ETTSVYEPRT ILKLLNDAIE SGSDQLPTNQ LNFVKYLKIF PLSKLENHKE AVHILLDEID DYKAATSYCN DVYQSDSTKG EELLLYLYS KLVSIYDSNR NSKLILNFLQ DHGSKLNSAE IYKNLPQDIS LYDIGRVVSQ LLKKHTSKMD ETRLEKALLQ V ELVATTYK LNERMSSYGV LSDSHKCPIC KKVISNFGTD SISWFTREGR NIITHYNCGK VLQERFNAKN EKSSRIKQKT LG EVINELN NK UniProtKB: Vacuolar morphogenesis protein 6 |
-Macromolecule #6: Vacuolar protein sorting-associated protein 41
| Macromolecule | Name: Vacuolar protein sorting-associated protein 41 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 116.530555 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTDNHQNDS VLDQQSGERT IDESNSISDE NNVDNKREDV NVTSPTKSVS CISQAENGVA SRTDESTITG SATDAETGDD DDDDDDDDD EDEDDEDEPP LLKYTRISQL PKNFFQRDSI SSCLFGDTFF AFGTHSGILH LTTCAFEPIK TIKCHRSSIL C INTDGKYF ...String: MTTDNHQNDS VLDQQSGERT IDESNSISDE NNVDNKREDV NVTSPTKSVS CISQAENGVA SRTDESTITG SATDAETGDD DDDDDDDDD EDEDDEDEPP LLKYTRISQL PKNFFQRDSI SSCLFGDTFF AFGTHSGILH LTTCAFEPIK TIKCHRSSIL C INTDGKYF ATGSIDGTVI IGSMDDPQNI TQYDFKRPIN SVALHSNFQA SRMFVSGGMA GDVVLSQRNW LGNRIDIVLN KK KKKKTRK DDLSSDMKGP IMGIYTMGDL ILWMDDDGIT FCDVPTRSQL LNIPFPSRIF NVQDVRPDLF RPHVHFLESD RVV IGWGSN IWLFKVSFTK DSNSIKSGDS NSQSNNMSHF NPTTNIGSLL SSAASSFRGT PDKKVELECH FTVSMLITGL ASFK DDQLL CLGFDIDIEE EATIDEDMKE GKNFSKRPEN LLAKGNAPEL KIVDLFNGDE IYNDEVIMKN YEKLSINDYH LGKHI DKTT PEYYLISSND AIRVQELSLK DHFDWFMERK QYYKAWKIGK YVIGSEERFS IGLKFLNSLV TKKDWGTLVD HLNIIF EET LNSLDSNSYD VTQNVLKEWA DIIEILITSG NIVEIAPLIP KKPALRKSVY DDVLHYFLAN DMINKFHEYI TKWDLKL FS VEDFEEELET RIEAASEPTA SSKEEGSNIT YRTELVHLYL KENKYTKAIP HLLKAKDLRA LTIIKIQNLL PQYLDQIV D IILLPYKGEI SHISKLSIFE IQTIFNKPID LLFENRHTIS VARIYEIFEH DCPKSFKKIL FCYLIKFLDT DDSFMISPY ENQLIELYSE YDRQSLLPFL QKHNNYNVES AIEVCSSKLG LYNELIYLWG KIGETKKALS LIIDELKNPQ LAIDFVKNWG DSELWEFMI NYSLDKPNFT KAILTCSDET SEIYLKVIRG MSDDLQIDNL QDIIKHIVQE NSLSLEVRDN ILVIINDETK K FANEFLKI RSQGKLFQVD ESDIEINDDL NGVLDYKDDD DKDYKDDDDK DYKDDDDK UniProtKB: Vacuolar protein sorting-associated protein 41 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.8 µm |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.4 Å / Resolution method: OTHER Details: Resolution of the consensus map of the lower part of the complex is used here Number images used: 244661 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.3.1) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.3.1) |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)