+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Non-muscle F-actin decorated with non-muscle tropomyosin 3.2 | |||||||||
 Map data Map data | Non-muscle F-actin decorated with non-muscle tropomyosin 3.2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | actin / tropomyosin / non-muscle / complex / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationActins signature 1. / Actin, conserved site / Actins signature 2. / Actin/actin-like conserved site / Actins and actin-related proteins signature. / Actin / Actin family / Actin / ATPase, nucleotide binding domain Similarity search - Domain/homology | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
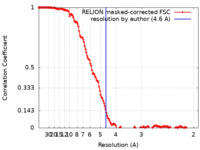
| Method | helical reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Selvaraj M / Kokate S / Kogan K / Kotila T / Kremneva E / Lappalainen P / Huiskonen JT | |||||||||
| Funding support |  Finland, 2 items Finland, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2023 Journal: Cell Rep / Year: 2023Title: Structural basis underlying specific biochemical activities of non-muscle tropomyosin isoforms. Authors: Muniyandi Selvaraj / Shrikant B Kokate / Gabriella Reggiano / Konstantin Kogan / Tommi Kotila / Elena Kremneva / Frank DiMaio / Pekka Lappalainen / Juha T Huiskonen /   Abstract: The actin cytoskeleton is critical for cell migration, morphogenesis, endocytosis, organelle dynamics, and cytokinesis. To support diverse cellular processes, actin filaments form a variety of ...The actin cytoskeleton is critical for cell migration, morphogenesis, endocytosis, organelle dynamics, and cytokinesis. To support diverse cellular processes, actin filaments form a variety of structures with specific architectures and dynamic properties. Key proteins specifying actin filaments are tropomyosins. Non-muscle cells express several functionally non-redundant tropomyosin isoforms, which differentially control the interactions of other proteins, including myosins and ADF/cofilin, with actin filaments. However, the underlying molecular mechanisms have remained elusive. By determining the cryogenic electron microscopy structures of actin filaments decorated by two functionally distinct non-muscle tropomyosin isoforms, Tpm1.6 and Tpm3.2, we reveal that actin filament conformation remains unaffected upon binding. However, Tpm1.6 and Tpm3.2 follow different paths along the actin filament major groove, providing an explanation for their incapability to co-polymerize on actin filaments. We also elucidate the molecular basis underlying specific roles of Tpm1.6 and Tpm3.2 in myosin II activation and protecting actin filaments from ADF/cofilin-catalyzed severing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14958.map.gz emd_14958.map.gz | 228.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14958-v30.xml emd-14958-v30.xml emd-14958.xml emd-14958.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14958_fsc.xml emd_14958_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14958.png emd_14958.png | 76.5 KB | ||
| Masks |  emd_14958_msk_1.map emd_14958_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14958.cif.gz emd-14958.cif.gz | 6 KB | ||
| Others |  emd_14958_half_map_1.map.gz emd_14958_half_map_1.map.gz emd_14958_half_map_2.map.gz emd_14958_half_map_2.map.gz | 194 MB 194 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14958 http://ftp.pdbj.org/pub/emdb/structures/EMD-14958 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14958 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14958 | HTTPS FTP |
-Validation report
| Summary document |  emd_14958_validation.pdf.gz emd_14958_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14958_full_validation.pdf.gz emd_14958_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_14958_validation.xml.gz emd_14958_validation.xml.gz | 21.6 KB | Display | |
| Data in CIF |  emd_14958_validation.cif.gz emd_14958_validation.cif.gz | 28.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14958 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14958 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14958 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14958 | HTTPS FTP |
-Related structure data
| Related structure data |  7ztdMC  7ztcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14958.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14958.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Non-muscle F-actin decorated with non-muscle tropomyosin 3.2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14958_msk_1.map emd_14958_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Halfmap 1
| File | emd_14958_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Halfmap 2
| File | emd_14958_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Non-muscle F-actin decorated with non-muscle tropomyosin 3.2
| Entire | Name: Non-muscle F-actin decorated with non-muscle tropomyosin 3.2 |
|---|---|
| Components |
|
-Supramolecule #1: Non-muscle F-actin decorated with non-muscle tropomyosin 3.2
| Supramolecule | Name: Non-muscle F-actin decorated with non-muscle tropomyosin 3.2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Beta actin (non-muscle)
| Supramolecule | Name: Beta actin (non-muscle) / type: organelle_or_cellular_component / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: Platelets Homo sapiens (human) / Tissue: Platelets |
-Supramolecule #3: Non-muscle tropomyosin 3.2
| Supramolecule | Name: Non-muscle tropomyosin 3.2 / type: organelle_or_cellular_component / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: actin, cytoplasmic 1
| Macromolecule | Name: actin, cytoplasmic 1 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: Platelets Homo sapiens (human) / Tissue: Platelets |
| Molecular weight | Theoretical: 41.78266 KDa |
| Sequence | String: MDDDIAALVV DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IEHGIVTNWD DMEKIWHHT FYNELRVAPE EHPVLLTEAP LNPKANREKM TQIMFETFNT PAMYVAIQAV LSLYASGRTT GIVMDSGDGV T HTVPIYEG ...String: MDDDIAALVV DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IEHGIVTNWD DMEKIWHHT FYNELRVAPE EHPVLLTEAP LNPKANREKM TQIMFETFNT PAMYVAIQAV LSLYASGRTT GIVMDSGDGV T HTVPIYEG YALPHAILRL DLAGRDLTDY LMKILTERGY SFTTTAEREI VRDIKEKLCY VALDFEQEMA TAASSSSLEK SY ELPDGQV ITIGNERFRC PEALFQPSFL GMESCGIHET TFNSIMKCDV DIRKDLYANT VLSGGTTMYP GIADRMQKEI TAL APSTMK IKIIAPPERK YSVWIGGSIL ASLSTFQQMW ISKQEYDESG PSIVHRKCF UniProtKB: Actin, cytoplasmic 1 |
-Macromolecule #2: Non-muscle tropomyosin 3.2
| Macromolecule | Name: Non-muscle tropomyosin 3.2 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.507176 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 8 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Number real images: 1250 / Average exposure time: 45.0 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)