+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1485 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The archaeal DNA Ligase-PCNA-DNA complex | |||||||||
 Map data Map data | This is a map of DNA ligase-PCNA-DNA complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Pyrococcus furiosus (archaea) / synthetic construct (others) Pyrococcus furiosus (archaea) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 17.0 Å | |||||||||
 Authors Authors | Mayanagi K / Kiyonari S / Ishino Y / Shirai T / Morikawa K | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2009 Journal: Proc Natl Acad Sci U S A / Year: 2009Title: Mechanism of replication machinery assembly as revealed by the DNA ligase-PCNA-DNA complex architecture. Authors: Kouta Mayanagi / Shinichi Kiyonari / Mihoko Saito / Tsuyoshi Shirai / Yoshizumi Ishino / Kosuke Morikawa /  Abstract: The 3D structure of the ternary complex, consisting of DNA ligase, the proliferating cell nuclear antigen (PCNA) clamp, and DNA, was investigated by single-particle analysis. This report presents the ...The 3D structure of the ternary complex, consisting of DNA ligase, the proliferating cell nuclear antigen (PCNA) clamp, and DNA, was investigated by single-particle analysis. This report presents the structural view, where the crescent-shaped DNA ligase with 3 distinct domains surrounds the central DNA duplex, encircled by the closed PCNA ring, thus forming a double-layer structure with dual contacts between the 2 proteins. The relative orientations of the DNA ligase domains, which remarkably differ from those of the known crystal structures, suggest that a large domain rearrangement occurs upon ternary complex formation. A second contact was found between the PCNA ring and the middle adenylation domain of the DNA ligase. Notably, the map revealed a substantial DNA tilt from the PCNA ring axis. This structure allows us to propose a switching mechanism for the replication factors operating on the PCNA ring. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1485.map.gz emd_1485.map.gz | 209.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1485-v30.xml emd-1485-v30.xml emd-1485.xml emd-1485.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| Images |  1485.png 1485.png | 286 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1485 http://ftp.pdbj.org/pub/emdb/structures/EMD-1485 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1485 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1485 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1485.map.gz / Format: CCP4 / Size: 670.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1485.map.gz / Format: CCP4 / Size: 670.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of DNA ligase-PCNA-DNA complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
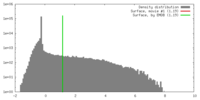
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Pyrococcus furiosus DNA Ligase bound to PCNA and DNA
| Entire | Name: Pyrococcus furiosus DNA Ligase bound to PCNA and DNA |
|---|---|
| Components |
|
-Supramolecule #1000: Pyrococcus furiosus DNA Ligase bound to PCNA and DNA
| Supramolecule | Name: Pyrococcus furiosus DNA Ligase bound to PCNA and DNA / type: sample / ID: 1000 / Details: The sample was monodisperse / Oligomeric state: Heteropentamer / Number unique components: 3 |
|---|---|
| Molecular weight | Theoretical: 160 KDa Method: One DNA Ligase binds to three PCNA and one nicked DNA |
-Macromolecule #1: Pyrococcus furiosus DNA Ligase
| Macromolecule | Name: Pyrococcus furiosus DNA Ligase / type: protein_or_peptide / ID: 1 / Name.synonym: DNA Ligase / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
-Macromolecule #2: Proliferating cell nuclear antigen
| Macromolecule | Name: Proliferating cell nuclear antigen / type: protein_or_peptide / ID: 2 / Name.synonym: PCNA / Number of copies: 3 / Oligomeric state: Trimeric / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Molecular weight | Theoretical: 28 KDa |
-Macromolecule #3: PCNA-nicked DNA
| Macromolecule | Name: PCNA-nicked DNA / type: dna / ID: 3 / Name.synonym: DNA / Classification: DNA / Structure: DOUBLE HELIX / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 6.5 / Details: 20 mM MES,50 mM NaCl,10 mM MgCl2,0.1 mM ATP |
| Staining | Type: NEGATIVE Details: The sample solution was applied to a copper grid supporting a continuous thin-carbon film, left for 1 min, then stained with 3 drops of 2% uranyl acetate, and air dried. |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1010 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 400,000 times magnification |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN / Number real images: 400 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 5.6 mm / Nominal magnification: 400000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: OTHER |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 17.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN,IMAGIC / Number images used: 19544 |
|---|
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Details | PDBEntryID_givenInChain. |
| Refinement | Space: REAL |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Details | PDBEntryID_givenInChain. |
| Refinement | Space: REAL |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera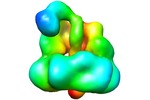








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)