[English] 日本語
 Yorodumi
Yorodumi- EMDB-14811: SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (Stat... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Focused Refinement | |||||||||
 Map data Map data | DeepEMhancer sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   | |||||||||
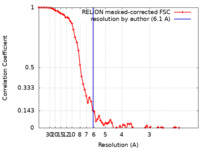
| Method | single particle reconstruction / cryo EM / Resolution: 6.1 Å | |||||||||
 Authors Authors | Hurdiss DL / Drulyte I | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Biotechnol / Year: 2022 Journal: Nat Biotechnol / Year: 2022Title: The trispecific DARPin ensovibep inhibits diverse SARS-CoV-2 variants. Authors: Sylvia Rothenberger / Daniel L Hurdiss / Marcel Walser / Francesca Malvezzi / Jennifer Mayor / Sarah Ryter / Hector Moreno / Nicole Liechti / Andreas Bosshart / Chloé Iss / Valérie Calabro ...Authors: Sylvia Rothenberger / Daniel L Hurdiss / Marcel Walser / Francesca Malvezzi / Jennifer Mayor / Sarah Ryter / Hector Moreno / Nicole Liechti / Andreas Bosshart / Chloé Iss / Valérie Calabro / Andreas Cornelius / Tanja Hospodarsch / Alexandra Neculcea / Thamar Looser / Anja Schlegel / Simon Fontaine / Denis Villemagne / Maria Paladino / Dieter Schiegg / Susanne Mangold / Christian Reichen / Filip Radom / Yvonne Kaufmann / Doris Schaible / Iris Schlegel / Christof Zitt / Gabriel Sigrist / Marcel Straumann / Julia Wolter / Marco Comby / Feyza Sacarcelik / Ieva Drulyte / Heyrhyoung Lyoo / Chunyan Wang / Wentao Li / Wenjuan Du / H Kaspar Binz / Rachel Herrup / Sabrina Lusvarghi / Sabari Nath Neerukonda / Russell Vassell / Wei Wang / Julia M Adler / Kathrin Eschke / Mariana Nascimento / Azza Abdelgawad / Achim D Gruber / Judith Bushe / Olivia Kershaw / Charles G Knutson / Kamal K Balavenkatraman / Krishnan Ramanathan / Emanuel Wyler / Luiz Gustavo Teixeira Alves / Seth Lewis / Randall Watson / Micha A Haeuptle / Alexander Zürcher / Keith M Dawson / Daniel Steiner / Carol D Weiss / Patrick Amstutz / Frank J M van Kuppeveld / Michael T Stumpp / Berend-Jan Bosch / Olivier Engler / Jakob Trimpert /     Abstract: The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants with potential resistance to existing drugs emphasizes the need for new therapeutic modalities with broad ...The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants with potential resistance to existing drugs emphasizes the need for new therapeutic modalities with broad variant activity. Here we show that ensovibep, a trispecific DARPin (designed ankyrin repeat protein) clinical candidate, can engage the three units of the spike protein trimer of SARS-CoV-2 and inhibit ACE2 binding with high potency, as revealed by cryo-electron microscopy analysis. The cooperative binding together with the complementarity of the three DARPin modules enable ensovibep to inhibit frequent SARS-CoV-2 variants, including Omicron sublineages BA.1 and BA.2. In Roborovski dwarf hamsters infected with SARS-CoV-2, ensovibep reduced fatality similarly to a standard-of-care monoclonal antibody (mAb) cocktail. When used as a single agent in viral passaging experiments in vitro, ensovibep reduced the emergence of escape mutations in a similar fashion to the same mAb cocktail. These results support further clinical evaluation of ensovibep as a broad variant alternative to existing targeted therapies for Coronavirus Disease 2019 (COVID-19). | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14811.map.gz emd_14811.map.gz | 27.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14811-v30.xml emd-14811-v30.xml emd-14811.xml emd-14811.xml | 26.8 KB 26.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14811_fsc.xml emd_14811_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14811.png emd_14811.png | 76.7 KB | ||
| Others |  emd_14811_additional_1.map.gz emd_14811_additional_1.map.gz emd_14811_additional_2.map.gz emd_14811_additional_2.map.gz emd_14811_half_map_1.map.gz emd_14811_half_map_1.map.gz emd_14811_half_map_2.map.gz emd_14811_half_map_2.map.gz | 20.8 MB 23.3 MB 23.5 MB 23.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14811 http://ftp.pdbj.org/pub/emdb/structures/EMD-14811 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14811 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14811 | HTTPS FTP |
-Validation report
| Summary document |  emd_14811_validation.pdf.gz emd_14811_validation.pdf.gz | 585.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14811_full_validation.pdf.gz emd_14811_full_validation.pdf.gz | 584.8 KB | Display | |
| Data in XML |  emd_14811_validation.xml.gz emd_14811_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_14811_validation.cif.gz emd_14811_validation.cif.gz | 17.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14811 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14811 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14811 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14811 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14811.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14811.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
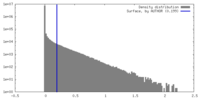
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Local resolution filtered map
| File | emd_14811_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
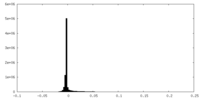
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_14811_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
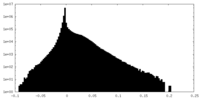
| Density Histograms |
-Half map: Half map 1
| File | emd_14811_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
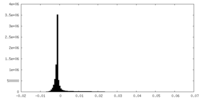
| Density Histograms |
-Half map: Half map 2
| File | emd_14811_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2
| Entire | Name: SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2
| Supramolecule | Name: SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 464 KDa |
-Supramolecule #2: SARS-CoV-2 spike glycoprotein
| Supramolecule | Name: SARS-CoV-2 spike glycoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Strain: Severe acute respiratory syndrome coronavirus 2 (Wuhan-Hu-1) |
-Supramolecule #3: monovalent DARPin R2
| Supramolecule | Name: monovalent DARPin R2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: SARS-CoV-2 spike glycoprotein
| Macromolecule | Name: SARS-CoV-2 spike glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYT NSFTRGVYYP D KVFRSSVL HSTQDLFLPF FS NVTWFHA IHVSGTNGTK RFD NPVLPF NDGVYFASTE KSNI IRGWI FGTTLDSKTQ SLLIV NNAT NVVIKVCEFQ FCNDPF LGV YYHKNNKSWM ESEFRVY SS ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYT NSFTRGVYYP D KVFRSSVL HSTQDLFLPF FS NVTWFHA IHVSGTNGTK RFD NPVLPF NDGVYFASTE KSNI IRGWI FGTTLDSKTQ SLLIV NNAT NVVIKVCEFQ FCNDPF LGV YYHKNNKSWM ESEFRVY SS ANNCTFEYVS QPFLMDLE G KQGNFKNLRE FVFKNIDGY FKIYSKHTPI NLVRDLPQGF SALEPLVDL PIGINITRFQ T LLALHRSY LTPGDSSSGW TA GAAAYYV GYLQPRTFLL KYN ENGTIT DAVDCALDPL SETK CTLKS FTVEKGIYQT SNFRV QPTE SIVRFPNITN LCPFGE VFN ATRFASVYAW NRKRISN CV ADYSVLYNSA SFSTFKCY G VSPTKLNDLC FTNVYADSF VIRGDEVRQI APGQTGKIAD YNYKLPDDF TGCVIAWNSN N LDSKVGGN YNYLYRLFRK SN LKPFERD ISTEIYQAGS TPC NGVEGF NCYFPLQSYG FQPT NGVGY QPYRVVVLSF ELLHA PATV CGPKKSTNLV KNKCVN FNF NGLTGTGVLT ESNKKFL PF QQFGRDIADT TDAVRDPQ T LEILDITPCS FGGVSVITP GTNTSNQVAV LYQDVNCTEV PVAIHADQL TPTWRVYSTG S NVFQTRAG CLIGAEHVNN SY ECDIPIG AGICASYQTQ TNS PAAARS VASQSIIAYT MSLG AENSV AYSNNSIAIP TNFTI SVTT EILPVSMTKT SVDCTM YIC GDSTECSNLL LQYGSFC TQ LNRALTGIAV EQDKNTQE V FAQVKQIYKT PPIKDFGGF NFSQILPDPS KPSKRSFIED LLFNKVTLA DAGFIKQYGD C LGDIAARD LICAQKFNGL TV LPPLLTD EMIAQYTSAL LAG TITSGW TFGAGAALQI PFAM QMAYR FNGIGVTQNV LYENQ KLIA NQFNSAIGKI QDSLSS TAS ALGKLQDVVN QNAQALN TL VKQLSSNFGA ISSVLNDI L SRLDPPEAEV QIDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAATKM SECVLGQSKR V DFCGKGYH LMSFPQSAPH GV VFLHVTY VPAQEKNFTT APA ICHDGK AHFPREGVFV SNGT HWFVT QRNFYEPQII TTDNT FVSG NCDVVIGIVN NTVYDP LQP ELDSFKEELD KYFKNHT SP DVDLGDISGI NASVVNIQ K EIDRLNEVAK NLNESLIDL LIKGSGYIPE APRDGQAYVR KDGEWVLLS TFLIKLVPRG S LEWSHPQF EK |
-Macromolecule #2: monovalent DARPin R2
| Macromolecule | Name: monovalent DARPin R2 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism:  |
| Sequence | String: MRGSHHHHHH GSDLGKKLLQ AARAGQLDEV RELLKAGADV NAKDREGKTP LHVAAQEGHL EIVEVLLKAG ADVNAKDVWG RTPLHLAAWI GHLEIVEVLL KAGADVNAKD VSGATPLHAA ALHGHLEIVE VLLNAGADVN AQDKSGKTPA DLAARAGHQD IAEVLQKAA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | Data collected with a 30 degree stage tilt. |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 3024 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.25 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)