[English] 日本語
 Yorodumi
Yorodumi- EMDB-14770: Late assembly intermediate of the proximal proton pumping module ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Late assembly intermediate of the proximal proton pumping module of complex I with assembly factor NDUFAF1 | |||||||||
 Map data Map data | late assembly intermediate of the proximal proton pumping module of complex I with assemble factor NDUFAF1 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Yarrowia lipolytica (yeast) Yarrowia lipolytica (yeast) | |||||||||
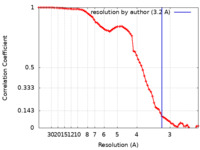
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Schiller J / Laube E / Vonck J / Zickermann V | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Insights into complex I assembly: Function of NDUFAF1 and a link with cardiolipin remodeling. Authors: Jonathan Schiller / Eike Laube / Ilka Wittig / Werner Kühlbrandt / Janet Vonck / Volker Zickermann /  Abstract: Respiratory complex I is a ~1-MDa proton pump in mitochondria. Its structure has been revealed in great detail, but the structural basis of its assembly, in humans involving at least 15 assembly ...Respiratory complex I is a ~1-MDa proton pump in mitochondria. Its structure has been revealed in great detail, but the structural basis of its assembly, in humans involving at least 15 assembly factors, is essentially unknown. We determined cryo-electron microscopy structures of assembly intermediates associated with assembly factor NDUFAF1 in a yeast model system. Subunits ND2 and NDUFC2 together with assembly factors NDUFAF1 and CIA84 form the nucleation point of the NDUFAF1-dependent assembly pathway. Unexpectedly, the cardiolipin remodeling enzyme tafazzin is an integral component of this core complex. In a later intermediate, all 12 subunits of the proximal proton pump module have assembled. NDUFAF1 locks the central ND3 subunit in an assembly-competent conformation, and major rearrangements of central subunits are required for complex I maturation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14770.map.gz emd_14770.map.gz | 32.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14770-v30.xml emd-14770-v30.xml emd-14770.xml emd-14770.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14770_fsc.xml emd_14770_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14770.png emd_14770.png | 147.6 KB | ||
| Masks |  emd_14770_msk_1.map emd_14770_msk_1.map | 34.3 MB |  Mask map Mask map | |
| Others |  emd_14770_additional_1.map.gz emd_14770_additional_1.map.gz emd_14770_half_map_1.map.gz emd_14770_half_map_1.map.gz emd_14770_half_map_2.map.gz emd_14770_half_map_2.map.gz | 6 MB 31.9 MB 31.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14770 http://ftp.pdbj.org/pub/emdb/structures/EMD-14770 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14770 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14770 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14770.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14770.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | late assembly intermediate of the proximal proton pumping module of complex I with assemble factor NDUFAF1 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.20721 Å | ||||||||||||||||||||||||||||||||||||
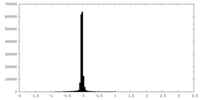
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14770_msk_1.map emd_14770_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
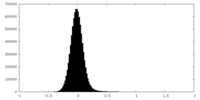
| Density Histograms |
-Additional map: map after density modification using Phenix
| File | emd_14770_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map after density modification using Phenix | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_14770_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14770_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : late Pp module assembly intermediate of complex I associated with...
| Entire | Name: late Pp module assembly intermediate of complex I associated with assembly factor NDUFAF1. |
|---|---|
| Components |
|
-Supramolecule #1: late Pp module assembly intermediate of complex I associated with...
| Supramolecule | Name: late Pp module assembly intermediate of complex I associated with assembly factor NDUFAF1. type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#14 |
|---|---|
| Source (natural) | Organism:  Yarrowia lipolytica (yeast) Yarrowia lipolytica (yeast) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 276 K / Instrument: FEI VITROBOT MARK IV |
| Details | The sample was a mixture of assembly intermediates associated with the assembly factor NDUFAF1 |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 30 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 6229 / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)