+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Early Pp module assembly intermediate of complex I | |||||||||
 Map data Map data | early Pp module assembly intermediate of complex I with assembly factors NDUFAF1 and CIA84 and cardiolipin remodeling enzyme tafazzin | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | assembly respiratory chain membrane protein mitochondria / ELECTRON TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationNADH dehydrogenase complex assembly / cardiolipin acyl-chain remodeling / 1-acylglycerophosphocholine O-acyltransferase activity / inner mitochondrial membrane organization / mitochondrial respirasome assembly / phospholipid biosynthetic process / NADH:ubiquinone reductase (H+-translocating) / mitochondrial ATP synthesis coupled electron transport / mitochondrial electron transport, NADH to ubiquinone / mitochondrial membrane ...NADH dehydrogenase complex assembly / cardiolipin acyl-chain remodeling / 1-acylglycerophosphocholine O-acyltransferase activity / inner mitochondrial membrane organization / mitochondrial respirasome assembly / phospholipid biosynthetic process / NADH:ubiquinone reductase (H+-translocating) / mitochondrial ATP synthesis coupled electron transport / mitochondrial electron transport, NADH to ubiquinone / mitochondrial membrane / unfolded protein binding / oxidoreductase activity / mitochondrial outer membrane / mitochondrial inner membrane / mitochondrion / membrane Similarity search - Function | |||||||||
| Biological species |  Yarrowia lipolytica (yeast) Yarrowia lipolytica (yeast) | |||||||||
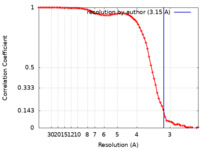
| Method | single particle reconstruction / cryo EM / Resolution: 3.15 Å | |||||||||
 Authors Authors | Schiller J / Laube E / Vonck J / Zickermann V | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Insights into complex I assembly: Function of NDUFAF1 and a link with cardiolipin remodeling. Authors: Jonathan Schiller / Eike Laube / Ilka Wittig / Werner Kühlbrandt / Janet Vonck / Volker Zickermann /  Abstract: Respiratory complex I is a ~1-MDa proton pump in mitochondria. Its structure has been revealed in great detail, but the structural basis of its assembly, in humans involving at least 15 assembly ...Respiratory complex I is a ~1-MDa proton pump in mitochondria. Its structure has been revealed in great detail, but the structural basis of its assembly, in humans involving at least 15 assembly factors, is essentially unknown. We determined cryo-electron microscopy structures of assembly intermediates associated with assembly factor NDUFAF1 in a yeast model system. Subunits ND2 and NDUFC2 together with assembly factors NDUFAF1 and CIA84 form the nucleation point of the NDUFAF1-dependent assembly pathway. Unexpectedly, the cardiolipin remodeling enzyme tafazzin is an integral component of this core complex. In a later intermediate, all 12 subunits of the proximal proton pump module have assembled. NDUFAF1 locks the central ND3 subunit in an assembly-competent conformation, and major rearrangements of central subunits are required for complex I maturation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14765.map.gz emd_14765.map.gz | 32.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14765-v30.xml emd-14765-v30.xml emd-14765.xml emd-14765.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14765_fsc.xml emd_14765_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14765.png emd_14765.png | 136.2 KB | ||
| Filedesc metadata |  emd-14765.cif.gz emd-14765.cif.gz | 7.7 KB | ||
| Others |  emd_14765_half_map_1.map.gz emd_14765_half_map_1.map.gz emd_14765_half_map_2.map.gz emd_14765_half_map_2.map.gz | 31.8 MB 31.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14765 http://ftp.pdbj.org/pub/emdb/structures/EMD-14765 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14765 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14765 | HTTPS FTP |
-Related structure data
| Related structure data |  7zkqMC  7zkpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14765.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14765.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | early Pp module assembly intermediate of complex I with assembly factors NDUFAF1 and CIA84 and cardiolipin remodeling enzyme tafazzin | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.20721 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14765_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14765_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Early Pp module assembly intermediate of complex I with assembly ...
| Entire | Name: Early Pp module assembly intermediate of complex I with assembly factors NDUFAF1 and CIA84 and cardiolipin remodeling enzyme tafazzin. |
|---|---|
| Components |
|
-Supramolecule #1: Early Pp module assembly intermediate of complex I with assembly ...
| Supramolecule | Name: Early Pp module assembly intermediate of complex I with assembly factors NDUFAF1 and CIA84 and cardiolipin remodeling enzyme tafazzin. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Yarrowia lipolytica (yeast) Yarrowia lipolytica (yeast) |
-Macromolecule #1: NADH dehydrogenase subunit 2
| Macromolecule | Name: NADH dehydrogenase subunit 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: NADH:ubiquinone reductase (H+-translocating) |
|---|---|
| Source (natural) | Organism:  Yarrowia lipolytica (yeast) Yarrowia lipolytica (yeast) |
| Molecular weight | Theoretical: 53.381367 KDa |
| Sequence | String: (FME)LILAIISLI TFVSMSKLSD NRAIIRLINI YLILVLVLDS FLYLLFLNNQ TYTVMGELLI FNSFTFYIDM LIYFIM IVI SSLYGYNLYN NNLYKTLFEP KKELIILFLI NILGALLIVH SNDFITLFVA IELQSYSIYL ITAIYNSSYK ASKASML YF FMGGILSILI ...String: (FME)LILAIISLI TFVSMSKLSD NRAIIRLINI YLILVLVLDS FLYLLFLNNQ TYTVMGELLI FNSFTFYIDM LIYFIM IVI SSLYGYNLYN NNLYKTLFEP KKELIILFLI NILGALLIVH SNDFITLFVA IELQSYSIYL ITAIYNSSYK ASKASML YF FMGGILSILI AYSINTYYSV LNSYTLHSLD SLIINTLDLN LILIALSLGL LFKIGIAPLH KWLISIYENT PILITIYI S LIPKISILSY LVLSNISINS LVISILAILT LLVGSVGGLL QIKIKRLLAF SGLTNAGYMM LLLLLNNNEF SYLYYITQY SISHLAIFMI IIFSIYYINY INNQYNPIIY VNQLKGLIHD NAYLVLSMAI VVFSFIGIPP LLGFFGKLNI LMSILNNGYY FISIVLIVA SLISALYYLY LLNVSIQDKN NILINSNETV SSVLSYILSS LIILITFGFI YNSLIIDIFN VYFN UniProtKB: NADH-ubiquinone oxidoreductase chain 2 |
-Macromolecule #2: Subunit NEBM of NADH:Ubiquinone Oxidoreductase (Complex I)
| Macromolecule | Name: Subunit NEBM of NADH:Ubiquinone Oxidoreductase (Complex I) type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yarrowia lipolytica (yeast) Yarrowia lipolytica (yeast) |
| Molecular weight | Theoretical: 8.059366 KDa |
| Sequence | String: MALFTSLVGA SGLGFATKFL SNKIRLKPAG YYPLGYVFSG VAWAGLGLVL HNVHQHSLEV LEKKKTALSE QRTE UniProtKB: Uncharacterized protein |
-Macromolecule #3: Complex I intermediate-associated protein 30-domain-containing protein
| Macromolecule | Name: Complex I intermediate-associated protein 30-domain-containing protein type: protein_or_peptide / ID: 3 / Details: C-terminal strep tag / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yarrowia lipolytica (yeast) Yarrowia lipolytica (yeast) |
| Molecular weight | Theoretical: 31.894016 KDa |
| Recombinant expression | Organism:  Yarrowia lipolytica (yeast) Yarrowia lipolytica (yeast) |
| Sequence | String: MSANPAIVRP TETTEQVLVN FTKPNSLETV LTKCDEELGG YSTVNLALER PTTGKPYGRF FGNLSLDLPK DNKMVTRSGF AMFRTLDQP SSMFKTNAWN WEQYRHLELR VRGDRRKYFV NVQSATPLAS DLYQHRLFIQ TPGEWETVVI PIDDFILTNK G VVQEQMAM ...String: MSANPAIVRP TETTEQVLVN FTKPNSLETV LTKCDEELGG YSTVNLALER PTTGKPYGRF FGNLSLDLPK DNKMVTRSGF AMFRTLDQP SSMFKTNAWN WEQYRHLELR VRGDRRKYFV NVQSATPLAS DLYQHRLFIQ TPGEWETVVI PIDDFILTNK G VVQEQMAM DTANVYTVGI GLIDRQYGPY NLDIEYIKAV AHPPLEFKPK KEYEVEKETI LLTPGQPMEL GKGKVKELEE NL YFQGAEA AAKEAAAKAW SHPQFEKGGG SGGGSGGSAW SHPQFEK UniProtKB: Complex I intermediate-associated protein 30-domain-containing protein |
-Macromolecule #4: complex I assembly factor CIA84
| Macromolecule | Name: complex I assembly factor CIA84 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yarrowia lipolytica (yeast) Yarrowia lipolytica (yeast) |
| Molecular weight | Theoretical: 97.098469 KDa |
| Sequence | String: MPKNALLRSA RQVAISRVFA TSRASHVVSH APILASVRPR SNPAPYRRNF SSSRALRNDY GLDTAERSLK ESLVPFNGAP VDRKVVRDQ LMELISVSPG QVFPISVIPV VKSAYYELFR ENERVLSAGD TKTLFGAVAG NNPEDVQDLP FVLAVYHQAE Q AAETNRDS ...String: MPKNALLRSA RQVAISRVFA TSRASHVVSH APILASVRPR SNPAPYRRNF SSSRALRNDY GLDTAERSLK ESLVPFNGAP VDRKVVRDQ LMELISVSPG QVFPISVIPV VKSAYYELFR ENERVLSAGD TKTLFGAVAG NNPEDVQDLP FVLAVYHQAE Q AAETNRDS RDNILLLGKY FLFQDRLDNF WKLLEAQIKT HDDVDAGFVK QLLELISVDP HLTLGNVARV LQLKTDNHVS SS DELRNAL SATLEQLYYK ENEGSEFFLS LVENHILDSK DFTPSDSVVA MILNTCVNEG REDLGQSVLR NVVSRVGNLS PGQ EDPQNC WGFWSSVAMD LHGSKTDVKA FISRLEALPH RTKATWDILI RYAVFKADLA GRNDLLQVRA LLAEMQKVGF EPDA ETYFD AYRSSKSIKP DVVHLFEAEL DIEKDTSIFA IEMDKALKNH DTLEALSIFY ESFEQGAQWE NKRLHMEAMT ELLIQ YAGL NDTSVADILQ LVQRIEPICA QGRIPYSAET AIAQNVLQRH SDTANFYTFM NRQYGNTADK VTKQDPQIRP HTYQVI HDY IYSCESERAD LAWEMYGLLH KFYVVPFADY YKAIKFFAQD VKRQDYALLT FQQIRKNHDL HGQPAATSEM VAFLFHE FA KTKYKRGIKR LHEVVALETS FDVNRDVLNE MMAAYVSVED LNRVQDCWAQ LQQLPPSIGA NNRSVDVLLS YFKDNIHY T ERTWQGIPEF GLLPTLENYE QYLINNCRTG NYRRALEITK NMEIDSGLKP TAKIIAAVYN YTFTEQRKLE VEQWAEKAH PEMWLELKEG DKLKSLCLPA NSDNDNVESL LKQASADMDE EMSGGIVKVE SV UniProtKB: Mitochondrial group I intron splicing factor CCM1 |
-Macromolecule #5: Tafazzin family protein
| Macromolecule | Name: Tafazzin family protein / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yarrowia lipolytica (yeast) / Strain: CLIB 122 / E 150 Yarrowia lipolytica (yeast) / Strain: CLIB 122 / E 150 |
| Molecular weight | Theoretical: 42.835367 KDa |
| Sequence | String: MSFRNVSLRG SQLLGKLDSR GWGWYVAKKW NIGLVYTMCK VFLRCKKVDI KGLDNLLEAH RQARLEGRGL LTVMNHTSVL DDPVVWGML PNDNGWIPYL MRWATGAKDI CYKNKLYSLF FGAGQVLPIT RFGIGGPFQP GMDMCVRLLN PNNKIKYSAK Y TPYLVHTN ...String: MSFRNVSLRG SQLLGKLDSR GWGWYVAKKW NIGLVYTMCK VFLRCKKVDI KGLDNLLEAH RQARLEGRGL LTVMNHTSVL DDPVVWGML PNDNGWIPYL MRWATGAKDI CYKNKLYSLF FGAGQVLPIT RFGIGGPFQP GMDMCVRLLN PNNKIKYSAK Y TPYLVHTN ATSYPFWRES NWVHFFPEGY VHQALEPHEG TMRYFRWGTS RAVLEPVTPP IIVPMFSHGL QKVFQEIPKG YE MEGNNTN KDRTISIRIG EPISETTVAG FRNEWINLCH KENVGLNAET MPDVLKNGQE AKDLRSKVAA YLREEVEKLR LTV PNMNPE LPEFKEPEFW SDIDKVHKGV YNHRGKVRML RNPTKGLIEV VEANKD UniProtKB: Tafazzin family protein |
-Macromolecule #6: CARDIOLIPIN
| Macromolecule | Name: CARDIOLIPIN / type: ligand / ID: 6 / Number of copies: 2 / Formula: CDL |
|---|---|
| Molecular weight | Theoretical: 1.464043 KDa |
| Chemical component information |  ChemComp-CDL: |
-Macromolecule #7: Lauryl Maltose Neopentyl Glycol
| Macromolecule | Name: Lauryl Maltose Neopentyl Glycol / type: ligand / ID: 7 / Number of copies: 1 / Formula: LMN |
|---|---|
| Molecular weight | Theoretical: 1.005188 KDa |
| Chemical component information |  ChemComp-AV0: |
-Macromolecule #8: DIUNDECYL PHOSPHATIDYL CHOLINE
| Macromolecule | Name: DIUNDECYL PHOSPHATIDYL CHOLINE / type: ligand / ID: 8 / Number of copies: 1 / Formula: PLC |
|---|---|
| Molecular weight | Theoretical: 622.834 Da |
| Chemical component information |  ChemComp-PLC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 276 K / Instrument: FEI VITROBOT MARK IV |
| Details | The sample was a mixture of assembly intermediates associated with the assembly factor NDUFAF1 |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 6229 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)