[English] 日本語
 Yorodumi
Yorodumi- EMDB-14726: Cryo-EM structure of ex vivo AA amyloid from renal tissue of a sh... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ex vivo AA amyloid from renal tissue of a short hair cat deceased in a shelter | |||||||||
 Map data Map data | autosharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AA amyloidosis / cat / Serum amyloid A / PROTEIN FIBRIL | |||||||||
| Function / homology | Serum amyloid A protein / : / Serum amyloid A protein / Serum amyloid A proteins signature. / Serum amyloid A proteins / high-density lipoprotein particle / acute-phase response / Serum amyloid A protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
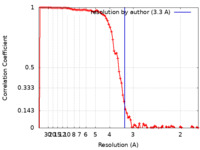
| Method | helical reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Schulte T / Chaves-Sanjuan A / Ricagno S | |||||||||
| Funding support |  Italy, 1 items Italy, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Cryo-EM structure of ex vivo fibrils associated with extreme AA amyloidosis prevalence in a cat shelter. Authors: Tim Schulte / Antonio Chaves-Sanjuan / Giulia Mazzini / Valentina Speranzini / Francesca Lavatelli / Filippo Ferri / Carlo Palizzotto / Maria Mazza / Paolo Milani / Mario Nuvolone / Anne- ...Authors: Tim Schulte / Antonio Chaves-Sanjuan / Giulia Mazzini / Valentina Speranzini / Francesca Lavatelli / Filippo Ferri / Carlo Palizzotto / Maria Mazza / Paolo Milani / Mario Nuvolone / Anne-Cathrine Vogt / Monique Vogel / Giovanni Palladini / Giampaolo Merlini / Martino Bolognesi / Silvia Ferro / Eric Zini / Stefano Ricagno /   Abstract: AA amyloidosis is a systemic disease characterized by deposition of misfolded serum amyloid A protein (SAA) into cross-β amyloid in multiple organs in humans and animals. AA amyloidosis occurs at ...AA amyloidosis is a systemic disease characterized by deposition of misfolded serum amyloid A protein (SAA) into cross-β amyloid in multiple organs in humans and animals. AA amyloidosis occurs at high SAA serum levels during chronic inflammation. Prion-like transmission was reported as possible cause of extreme AA amyloidosis prevalence in captive animals, e.g. 70% in cheetah and 57-73% in domestic short hair (DSH) cats kept in zoos and shelters, respectively. Herein, we present the 3.3 Å cryo-EM structure of AA amyloid extracted post-mortem from the kidney of a DSH cat with renal failure, deceased in a shelter with extreme disease prevalence. The structure reveals a cross-β architecture assembled from two 76-residue long proto-filaments. Despite >70% sequence homology to mouse and human SAA, the cat SAA variant adopts a distinct amyloid fold. Inclusion of an eight-residue insert unique to feline SAA contributes to increased amyloid stability. The presented feline AA amyloid structure is fully compatible with the 99% identical amino acid sequence of amyloid fragments of captive cheetah. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14726.map.gz emd_14726.map.gz | 53.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14726-v30.xml emd-14726-v30.xml emd-14726.xml emd-14726.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14726_fsc.xml emd_14726_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_14726.png emd_14726.png | 135.6 KB | ||
| Masks |  emd_14726_msk_1.map emd_14726_msk_1.map | 59.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14726.cif.gz emd-14726.cif.gz | 5.6 KB | ||
| Others |  emd_14726_additional_1.map.gz emd_14726_additional_1.map.gz emd_14726_half_map_1.map.gz emd_14726_half_map_1.map.gz emd_14726_half_map_2.map.gz emd_14726_half_map_2.map.gz | 44.1 MB 44.5 MB 44.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14726 http://ftp.pdbj.org/pub/emdb/structures/EMD-14726 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14726 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14726 | HTTPS FTP |
-Related structure data
| Related structure data |  7zh7MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14726.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14726.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | autosharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.889 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14726_msk_1.map emd_14726_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: map after 3D autorefine with gold-standard solvent-flattened fsc
| File | emd_14726_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map after 3D autorefine with gold-standard solvent-flattened fsc | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_14726_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_14726_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : AA amyloid extracted from kidney of deceased cat
| Entire | Name: AA amyloid extracted from kidney of deceased cat |
|---|---|
| Components |
|
-Supramolecule #1: AA amyloid extracted from kidney of deceased cat
| Supramolecule | Name: AA amyloid extracted from kidney of deceased cat / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Serum amyloid A protein
| Macromolecule | Name: Serum amyloid A protein / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.449766 KDa |
| Sequence | String: MKLFTGLVFC SLVLGVSSEW YSFLGEAAQG AWDMWRAYSD MREANYIGAD KYFHARGNYD AAQRGPGGAW AAKVISDARE NSQRVTDFF RHGNSGHGAE DSKADQEANE WGRSGKDPNH YRPEGLPDKY UniProtKB: Serum amyloid A protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. Details: glow discharged for 30s at 30mA using a GloQube system |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2652 / Average electron dose: 40.0 e/Å2 / Details: images takes for analysis |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 120000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 76.7 |
|---|---|
| Output model |  PDB-7zh7: |
 Movie
Movie Controller
Controller



 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)