+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1471 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Structural Basis of Membrane Invagination by F-BAR Domains | |||||||||
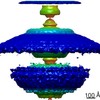
 Map data Map data | 3D reconstruction of a membrane tubule coated by a helical lattice of dimeric F-BAR modules from the human protein CIP4. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | F-BAR / FCH / PCH / CIP4 / FBP17 / Toca / Membrane Tubule. Human Protein. | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 17.0 Å | |||||||||
 Authors Authors | Frost A / Perera R / Roux A / Spasov K / Egelman E / De Camilli P / Unger V | |||||||||
 Citation Citation |  Journal: Cell / Year: 2008 Journal: Cell / Year: 2008Title: Structural basis of membrane invagination by F-BAR domains. Authors: Adam Frost / Rushika Perera / Aurélien Roux / Krasimir Spasov / Olivier Destaing / Edward H Egelman / Pietro De Camilli / Vinzenz M Unger /  Abstract: BAR superfamily domains shape membranes through poorly understood mechanisms. We solved structures of F-BAR modules bound to flat and curved bilayers using electron (cryo)microscopy. We show that ...BAR superfamily domains shape membranes through poorly understood mechanisms. We solved structures of F-BAR modules bound to flat and curved bilayers using electron (cryo)microscopy. We show that membrane tubules form when F-BARs polymerize into helical coats that are held together by lateral and tip-to-tip interactions. On gel-state membranes or after mutation of residues along the lateral interaction surface, F-BARs adsorb onto bilayers via surfaces other than their concave face. We conclude that membrane binding is separable from membrane bending, and that imposition of the module's concave surface forces fluid-phase bilayers to bend locally. Furthermore, exposure of the domain's lateral interaction surface through a change in orientation serves as the crucial trigger for assembly of the helical coat and propagation of bilayer bending. The geometric constraints and sequential assembly of the helical lattice explain how F-BAR and classical BAR domains segregate into distinct microdomains, and provide insight into the spatial regulation of membrane invagination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1471.map.gz emd_1471.map.gz | 62.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1471-v30.xml emd-1471-v30.xml emd-1471.xml emd-1471.xml | 8.3 KB 8.3 KB | Display Display |  EMDB header EMDB header |
| Images |  1471.gif 1471.gif | 79.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1471 http://ftp.pdbj.org/pub/emdb/structures/EMD-1471 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1471 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1471 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1471.map.gz / Format: CCP4 / Size: 238.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1471.map.gz / Format: CCP4 / Size: 238.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of a membrane tubule coated by a helical lattice of dimeric F-BAR modules from the human protein CIP4. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.28 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : membrane tubule coated with a helical lattice of dimer F-BAR modu...
| Entire | Name: membrane tubule coated with a helical lattice of dimer F-BAR modules from the human protein CIP4 |
|---|---|
| Components |
|
-Supramolecule #1000: membrane tubule coated with a helical lattice of dimer F-BAR modu...
| Supramolecule | Name: membrane tubule coated with a helical lattice of dimer F-BAR modules from the human protein CIP4 type: sample / ID: 1000 Details: membrane composed of phospholipids and cholesterol. recombinant protein Number unique components: 2 |
|---|
-Macromolecule #1: CIP4
| Macromolecule | Name: CIP4 / type: protein_or_peptide / ID: 1 / Name.synonym: CIP4 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Location in cell: plasmsa membrane and cytoplasm Homo sapiens (human) / synonym: Human / Location in cell: plasmsa membrane and cytoplasm |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | Jan 3, 2006 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder: eucentric, single tilt / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Axial symmetry: C2 (2 fold cyclic) Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 17.0 Å / Resolution method: OTHER / Software - Name: SPIDER, MRC, IHRSR |
|---|---|
| CTF correction | Details: ACE, image |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























