[English] 日本語
 Yorodumi
Yorodumi- EMDB-8569: In vivo structure of the Legionella pneumophila Dot/Icm type IV s... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8569 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
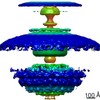
| Title | In vivo structure of the Legionella pneumophila Dot/Icm type IV secretion system core complex (aligning the inner membrane part) | |||||||||
 Map data Map data | Communication In vivo structure of the Legionella pneumophila Dot/Icm type IV secretion system core complex (aligning the inner membrane part) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 30.0 Å | |||||||||
 Authors Authors | Ghosal D / Chang YW / Jensen GJ | |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2017 Journal: EMBO Rep / Year: 2017Title: structure of the Dot/Icm type IV secretion system by electron cryotomography. Authors: Debnath Ghosal / Yi-Wei Chang / Kwangcheol C Jeong / Joseph P Vogel / Grant J Jensen /  Abstract: Type IV secretion systems (T4SSs) are large macromolecular machines that translocate protein and DNA and are involved in the pathogenesis of multiple human diseases. Here, using electron ...Type IV secretion systems (T4SSs) are large macromolecular machines that translocate protein and DNA and are involved in the pathogenesis of multiple human diseases. Here, using electron cryotomography (ECT), we report the structure of the Dot/Icm type IVB secretion system (T4BSS) utilized by the human pathogen This is the first structure of a type IVB secretion system, and also the first structure of any T4SS While the Dot/Icm system shares almost no sequence similarity with type IVA secretion systems (T4ASSs), its overall structure is seen here to be remarkably similar to previously reported T4ASS structures (those encoded by the R388 plasmid in and the cag pathogenicity island in ). This structural similarity suggests shared aspects of mechanism. However, compared to the negative-stain reconstruction of the purified T4ASS from the R388 plasmid, the Dot/Icm system is approximately twice as long and wide and exhibits several additional large densities, reflecting type-specific elaborations and potentially better structural preservation . | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8569.map.gz emd_8569.map.gz | 3.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8569-v30.xml emd-8569-v30.xml emd-8569.xml emd-8569.xml | 8.7 KB 8.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8569.png emd_8569.png | 28.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8569 http://ftp.pdbj.org/pub/emdb/structures/EMD-8569 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8569 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8569 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8569.map.gz / Format: CCP4 / Size: 4.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8569.map.gz / Format: CCP4 / Size: 4.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Communication In vivo structure of the Legionella pneumophila Dot/Icm type IV secretion system core complex (aligning the inner membrane part) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 8.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Legionella pneumophila Dot/Icm type IVB secretion system core com...
| Entire | Name: Legionella pneumophila Dot/Icm type IVB secretion system core complex (inner membrane complex) |
|---|---|
| Components |
|
-Supramolecule #1: Legionella pneumophila Dot/Icm type IVB secretion system core com...
| Supramolecule | Name: Legionella pneumophila Dot/Icm type IVB secretion system core complex (inner membrane complex) type: complex / ID: 1 / Parent: 0 Details: We used electron cryotomography and subtomogram averaging to visualize L. pneumophila Dot/Icm core complex directly in intact, frozen-hydrated bacteria cells. |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 6.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 30.0 Å / Resolution method: OTHER / Software - Name: IMOD, TOMO3D / Details: RESMAP / Number subtomograms used: 261 |
|---|---|
| Extraction | Number tomograms: 73 / Number images used: 261 / Software - Name: PEET |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















