[English] 日本語
 Yorodumi
Yorodumi- EMDB-14678: Cryo-EM structure of the human inward-rectifier potassium 2.1 cha... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human inward-rectifier potassium 2.1 channel (Kir2.1) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Potassium channel / Inward-rectifier channel / inward rectification / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationSensory perception of sour taste / Classical Kir channels / regulation of skeletal muscle contraction via regulation of action potential / relaxation of skeletal muscle / voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization / magnesium ion transport / membrane repolarization during action potential / Phase 4 - resting membrane potential / membrane repolarization during cardiac muscle cell action potential / regulation of membrane repolarization ...Sensory perception of sour taste / Classical Kir channels / regulation of skeletal muscle contraction via regulation of action potential / relaxation of skeletal muscle / voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization / magnesium ion transport / membrane repolarization during action potential / Phase 4 - resting membrane potential / membrane repolarization during cardiac muscle cell action potential / regulation of membrane repolarization / membrane depolarization during cardiac muscle cell action potential / regulation of resting membrane potential / regulation of monoatomic ion transmembrane transport / inward rectifier potassium channel activity / positive regulation of potassium ion transmembrane transport / cardiac muscle cell action potential involved in contraction / regulation of cardiac muscle cell contraction / relaxation of cardiac muscle / potassium ion import across plasma membrane / intracellular potassium ion homeostasis / regulation of heart rate by cardiac conduction / intercalated disc / phosphatidylinositol-4,5-bisphosphate binding / voltage-gated potassium channel complex / potassium ion transmembrane transport / T-tubule / cellular response to mechanical stimulus / potassium ion transport / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / protein homotetramerization / dendritic spine / postsynaptic membrane / neuronal cell body / glutamatergic synapse / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | ||||||||||||
 Authors Authors | Fernandes CAH / Venien-Bryan C / Fagnen C / Zuniga D | ||||||||||||
| Funding support |  France, 3 items France, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Cryo-electron microscopy unveils unique structural features of the human Kir2.1 channel. Authors: Carlos A H Fernandes / Dania Zuniga / Charline Fagnen / Valérie Kugler / Rosa Scala / Gérard Péhau-Arnaudet / Renaud Wagner / David Perahia / Saïd Bendahhou / Catherine Vénien-Bryan /  Abstract: We present the first structure of the human Kir2.1 channel containing both transmembrane domain (TMD) and cytoplasmic domain (CTD). Kir2.1 channels are strongly inward-rectifying potassium channels ...We present the first structure of the human Kir2.1 channel containing both transmembrane domain (TMD) and cytoplasmic domain (CTD). Kir2.1 channels are strongly inward-rectifying potassium channels that play a key role in maintaining resting membrane potential. Their gating is modulated by phosphatidylinositol 4,5-bisphosphate (PIP). Genetically inherited defects in Kir2.1 channels are responsible for several rare human diseases, including Andersen's syndrome. The structural analysis (cryo-electron microscopy), surface plasmon resonance, and electrophysiological experiments revealed a well-connected network of interactions between the PIP-binding site and the G-loop through residues R312 and H221. In addition, molecular dynamics simulations and normal mode analysis showed the intrinsic tendency of the CTD to tether to the TMD and a movement of the secondary anionic binding site to the membrane even without PIP. Our results revealed structural features unique to human Kir2.1 and provided insights into the connection between G-loop and gating and the pathological mechanisms associated with this channel. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14678.map.gz emd_14678.map.gz | 28.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14678-v30.xml emd-14678-v30.xml emd-14678.xml emd-14678.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14678_fsc.xml emd_14678_fsc.xml | 1.7 MB | Display |  FSC data file FSC data file |
| Images |  emd_14678.png emd_14678.png | 73.6 KB | ||
| Filedesc metadata |  emd-14678.cif.gz emd-14678.cif.gz | 6.9 KB | ||
| Others |  emd_14678_half_map_1.map.gz emd_14678_half_map_1.map.gz emd_14678_half_map_2.map.gz emd_14678_half_map_2.map.gz | 20.4 MB 20.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14678 http://ftp.pdbj.org/pub/emdb/structures/EMD-14678 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14678 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14678 | HTTPS FTP |
-Validation report
| Summary document |  emd_14678_validation.pdf.gz emd_14678_validation.pdf.gz | 819.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14678_full_validation.pdf.gz emd_14678_full_validation.pdf.gz | 819.2 KB | Display | |
| Data in XML |  emd_14678_validation.xml.gz emd_14678_validation.xml.gz | 10.9 KB | Display | |
| Data in CIF |  emd_14678_validation.cif.gz emd_14678_validation.cif.gz | 12.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14678 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14678 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14678 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14678 | HTTPS FTP |
-Related structure data
| Related structure data |  7zdzMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14678.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14678.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
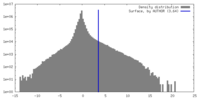
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14678_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
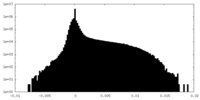
| Projections & Slices |
| ||||||||||||
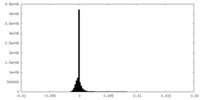
| Density Histograms |
-Half map: #1
| File | emd_14678_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
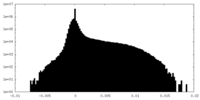
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human inward-rectifier potassium channel 2.1 (Kir2.1)
| Entire | Name: Human inward-rectifier potassium channel 2.1 (Kir2.1) |
|---|---|
| Components |
|
-Supramolecule #1: Human inward-rectifier potassium channel 2.1 (Kir2.1)
| Supramolecule | Name: Human inward-rectifier potassium channel 2.1 (Kir2.1) / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50 kDa/nm |
-Macromolecule #1: Inward rectifier potassium channel 2
| Macromolecule | Name: Inward rectifier potassium channel 2 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 48.344141 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MGSVRTNRYS IVSSEEDGMK LATMAVANGF GNGKSKVHTR QQCRSRFVKK DGHCNVQFIN VGEKGQRYLA DIFTTCVDIR WRWMLVIFC LAFVLSWLFF GCVFWLIALL HGDLDASKEG KACVSEVNSF TAAFLFSIET QTTIGYGFRC VTDECPIAVF M VVFQSIVG ...String: MGSVRTNRYS IVSSEEDGMK LATMAVANGF GNGKSKVHTR QQCRSRFVKK DGHCNVQFIN VGEKGQRYLA DIFTTCVDIR WRWMLVIFC LAFVLSWLFF GCVFWLIALL HGDLDASKEG KACVSEVNSF TAAFLFSIET QTTIGYGFRC VTDECPIAVF M VVFQSIVG CIIDAFIIGA VMAKMAKPKK RNETLVFSHN AVIAMRDGKL CLMWRVGNLR KSHLVEAHVR AQLLKSRITS EG EYIPLDQ IDINVGFDSG IDRIFLVSPI TIVHEIDEDS PLYDLSKQDI DNADFEIVVI LEGMVEATAM TTQCRSSYLA NEI LWGHRY EPVLFEEKHY YKVDYSRFHK TYEVPNTPLC SARDLAEKKY ILSNANSFCY ENEVALTSKE EDDSENGVPE STST DTPPD IDLHNQASVP LEPRPLRRES EI UniProtKB: Inward rectifier potassium channel 2 |
-Macromolecule #2: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #3: STRONTIUM ION
| Macromolecule | Name: STRONTIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: SR |
|---|---|
| Molecular weight | Theoretical: 87.62 Da |
| Chemical component information |  ChemComp-SR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 9895 / Average exposure time: 4.0 sec. / Average electron dose: 61.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Residue range: 188-367 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | For structural fitting, it was used dock-in-map (available at PHENIX) that uses both SSM and convolution-based shape searches to find a part of a map that is similar to a model. An initial in silico homology model of human Kir2.1 was generated using I-TASSER using the crystal structure of chicken Kir2.2 channel (PDB ID 3JYC) as a template. For building and refinement of the atomic model, the transmembrane domain (TMD, 55-184 region) of this in silico model was placed into the final sharpened cryo-EM map using the Dock in Map tool available in PHENIX. For the cytoplasmic domain (CTD; 188-367 region), the crystal structure of the CTD from mice Kir2.1 channel (PDB ID 1U4F) was placed into the final cryo-EM map using the same approach. Once the models were placed in the electron density, the loops that connect the two domains (185-187 region) and a N-terminal loop (41-54 region) absent in the in silico model were manually built using Coot. |
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 404.73 / Target criteria: Correlation coefficient |
| Output model |  PDB-7zdz: |
 Movie
Movie Controller
Controller




 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)