+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Complex I from Ovis aries at pH7.4, Closed state | |||||||||
 マップデータ マップデータ | Composite map | |||||||||
 試料 試料 |
| |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報myoblast migration involved in skeletal muscle regeneration / negative regulation of skeletal muscle satellite cell proliferation / : / : / oxidoreductase activity, acting on NAD(P)H / NADH dehydrogenase activity / positive regulation of macrophage chemotaxis / ubiquinone binding / electron transport coupled proton transport / acyl binding ...myoblast migration involved in skeletal muscle regeneration / negative regulation of skeletal muscle satellite cell proliferation / : / : / oxidoreductase activity, acting on NAD(P)H / NADH dehydrogenase activity / positive regulation of macrophage chemotaxis / ubiquinone binding / electron transport coupled proton transport / acyl binding / acyl carrier activity / apoptotic mitochondrial changes / NADH:ubiquinone reductase (H+-translocating) / mitochondrial ATP synthesis coupled electron transport / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / respiratory chain complex I / membrane => GO:0016020 / positive regulation of myoblast differentiation / NADH dehydrogenase (ubiquinone) activity / quinone binding / ATP metabolic process / ATP synthesis coupled electron transport / positive regulation of lamellipodium assembly / reactive oxygen species metabolic process / regulation of mitochondrial membrane potential / respiratory electron transport chain / transcription coregulator activity / electron transport chain / circadian rhythm / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / NAD binding / FMN binding / 4 iron, 4 sulfur cluster binding / response to oxidative stress / mitochondrial inner membrane / mitochondrial matrix / chromatin / protein-containing complex binding / positive regulation of transcription by RNA polymerase II / mitochondrion / metal ion binding / nucleus / membrane 類似検索 - 分子機能 | |||||||||
| 生物種 |   | |||||||||
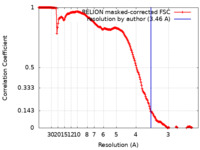
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.46 Å | |||||||||
 データ登録者 データ登録者 | Sazanov L / Petrova O | |||||||||
| 資金援助 | European Union, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2022 ジャーナル: Nature / 年: 2022タイトル: A universal coupling mechanism of respiratory complex I. 著者: Vladyslav Kravchuk / Olga Petrova / Domen Kampjut / Anna Wojciechowska-Bason / Zara Breese / Leonid Sazanov /    要旨: Complex I is the first enzyme in the respiratory chain, which is responsible for energy production in mitochondria and bacteria. Complex I couples the transfer of two electrons from NADH to quinone ...Complex I is the first enzyme in the respiratory chain, which is responsible for energy production in mitochondria and bacteria. Complex I couples the transfer of two electrons from NADH to quinone and the translocation of four protons across the membrane, but the coupling mechanism remains contentious. Here we present cryo-electron microscopy structures of Escherichia coli complex I (EcCI) in different redox states, including catalytic turnover. EcCI exists mostly in the open state, in which the quinone cavity is exposed to the cytosol, allowing access for water molecules, which enable quinone movements. Unlike the mammalian paralogues, EcCI can convert to the closed state only during turnover, showing that closed and open states are genuine turnover intermediates. The open-to-closed transition results in the tightly engulfed quinone cavity being connected to the central axis of the membrane arm, a source of substrate protons. Consistently, the proportion of the closed state increases with increasing pH. We propose a detailed but straightforward and robust mechanism comprising a 'domino effect' series of proton transfers and electrostatic interactions: the forward wave ('dominoes stacking') primes the pump, and the reverse wave ('dominoes falling') results in the ejection of all pumped protons from the distal subunit NuoL. This mechanism explains why protons exit exclusively from the NuoL subunit and is supported by our mutagenesis data. We contend that this is a universal coupling mechanism of complex I and related enzymes. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_14648.map.gz emd_14648.map.gz | 3.9 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-14648-v30.xml emd-14648-v30.xml emd-14648.xml emd-14648.xml | 57.5 KB 57.5 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_14648_fsc.xml emd_14648_fsc.xml | 17.8 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_14648.png emd_14648.png | 54.8 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14648 http://ftp.pdbj.org/pub/emdb/structures/EMD-14648 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14648 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14648 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_14648_validation.pdf.gz emd_14648_validation.pdf.gz | 402.4 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_14648_full_validation.pdf.gz emd_14648_full_validation.pdf.gz | 402 KB | 表示 | |
| XML形式データ |  emd_14648_validation.xml.gz emd_14648_validation.xml.gz | 12.2 KB | 表示 | |
| CIF形式データ |  emd_14648_validation.cif.gz emd_14648_validation.cif.gz | 17.7 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14648 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14648 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14648 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14648 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7zdhMC  7p61C  7p62C  7p63C  7p64C  7p69C  7p7cC  7p7eC  7p7jC  7p7kC  7p7lC  7p7mC  7z7rC  7z7sC  7z7tC  7z7vC  7z80C  7z83C  7z84C  7zc5C  7zciC  7zd6C  7zdjC  7zdmC  7zdpC  7zebC C: 同じ文献を引用 ( M: このマップから作成された原子モデル |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_14648.map.gz / 形式: CCP4 / 大きさ: 20.4 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_14648.map.gz / 形式: CCP4 / 大きさ: 20.4 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Composite map | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 これらの図は立方格子座標系で作成されたものです | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.22 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
+全体 : Complex I from Ovis aries at pH7.4, Closed state
+超分子 #1: Complex I from Ovis aries at pH7.4, Closed state
+分子 #1: NADH-ubiquinone oxidoreductase chain 3
+分子 #2: NADH-ubiquinone oxidoreductase chain 1
+分子 #3: NADH-ubiquinone oxidoreductase chain 6
+分子 #4: NADH-ubiquinone oxidoreductase chain 4L
+分子 #5: NADH-ubiquinone oxidoreductase chain 5
+分子 #6: NADH-ubiquinone oxidoreductase chain 4
+分子 #7: NADH-ubiquinone oxidoreductase chain 2
+分子 #8: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11
+分子 #9: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mito...
+分子 #10: Acyl carrier protein
+分子 #11: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8
+分子 #12: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10
+分子 #13: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mi...
+分子 #14: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5
+分子 #15: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 3
+分子 #16: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3
+分子 #17: NADH dehydrogenase [ubiquinone] 1 subunit C2
+分子 #18: NADH:ubiquinone oxidoreductase subunit B4
+分子 #19: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13
+分子 #20: Mitochondrial complex I, B17 subunit
+分子 #21: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7
+分子 #22: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9
+分子 #23: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2, mito...
+分子 #24: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mito...
+分子 #25: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mit...
+分子 #26: NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial
+分子 #27: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1
+分子 #28: Complex I-MWFE
+分子 #29: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial
+分子 #30: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial
+分子 #31: NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial
+分子 #32: NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial
+分子 #33: NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial
+分子 #34: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial
+分子 #35: Complex I-23kD
+分子 #36: NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial
+分子 #37: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial
+分子 #38: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial
+分子 #39: NADH:ubiquinone oxidoreductase subunit A9
+分子 #40: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2
+分子 #41: Mitochondrial complex I, B13 subunit
+分子 #42: NADH:ubiquinone oxidoreductase subunit A6
+分子 #43: Mitochondrial complex I, B14.5a subunit
+分子 #44: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12
+分子 #45: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+分子 #46: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+分子 #47: S-[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-b...
+分子 #48: ADENOSINE MONOPHOSPHATE
+分子 #49: MYRISTIC ACID
+分子 #50: IRON/SULFUR CLUSTER
+分子 #51: FLAVIN MONONUCLEOTIDE
+分子 #52: 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE
+分子 #53: FE2/S2 (INORGANIC) CLUSTER
+分子 #54: POTASSIUM ION
+分子 #55: ZINC ION
+分子 #56: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 3 mg/mL |
|---|---|
| 緩衝液 | pH: 7.4 |
| グリッド | モデル: Quantifoil / 材質: COPPER |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | TFS GLACIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: FEI FALCON III (4k x 4k) 平均露光時間: 3.3 sec. / 平均電子線量: 90.0 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 1.0 µm / 倍率(公称値): 120000 |
| 試料ステージ | ホルダー冷却材: NITROGEN |
 ムービー
ムービー コントローラー
コントローラー
























































































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)