+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Leishmania RNA virus 1 virion | |||||||||
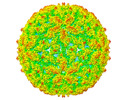
 Map data Map data | Virion structure | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | virion / full particle / VIRUS | |||||||||
| Function / homology | Totivirus coat / Totivirus coat protein / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Leishmania RNA virus 1 - 4 Leishmania RNA virus 1 - 4 | |||||||||
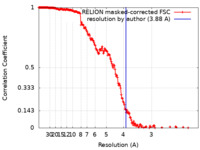
| Method | single particle reconstruction / cryo EM / Resolution: 3.88 Å | |||||||||
 Authors Authors | Prochazkova M / Plevka P | |||||||||
| Funding support |  Czech Republic, 1 items Czech Republic, 1 items
| |||||||||
 Citation Citation |  Journal: Virology / Year: 2022 Journal: Virology / Year: 2022Title: Virion structure of Leishmania RNA virus 1. Authors: Michaela Procházková / Tibor Füzik / Danyil Grybchuk / Vyacheslav Yurchenko / Pavel Plevka /  Abstract: The presence of Leishmania RNA virus 1 (LRV1) enables Leishmania protozoan parasites to cause more severe disease than the virus-free strains. The structure of LRV1 virus-like particles has been ...The presence of Leishmania RNA virus 1 (LRV1) enables Leishmania protozoan parasites to cause more severe disease than the virus-free strains. The structure of LRV1 virus-like particles has been determined previously, however, the structure of the LRV1 virion has not been characterized. Here we used cryo-electron microscopy and single-particle reconstruction to determine the structures of the LRV1 virion and empty particle isolated from Leishmania guyanensis to resolutions of 4.0 Å and 3.6 Å, respectively. The capsid of LRV1 is built from sixty dimers of capsid proteins organized with icosahedral symmetry. RNA genomes of totiviruses are replicated inside the virions by RNA polymerases expressed as C-terminal extensions of a sub-population of capsid proteins. Most of the virions probably contain one or two copies of the RNA polymerase, however, the location of the polymerase domains in LRV1 capsid could not be identified, indicating that it varies among particles. Importance. Every year over 200 000 people contract leishmaniasis and more than five hundred people die of the disease. The mucocutaneous form of leishmaniasis produces lesions that can destroy the mucous membranes of the nose, mouth, and throat. Leishmania parasites carrying Leishmania RNA virus 1 (LRV1) are predisposed to cause aggravated symptoms in the mucocutaneous form of leishmaniasis. Here, we present the structure of the LRV1 virion determined using cryo-electron microscopy. #1:  Journal: J Virol / Year: 2021 Journal: J Virol / Year: 2021Title: Capsid Structure of Leishmania RNA virus 1 Authors: Prochazkova M / Fuzik T / Grybchuk D / Falginella FL / Podesvova L / Yurchenko V / Vacha R / Plevka P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14566.map.gz emd_14566.map.gz | 59.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14566-v30.xml emd-14566-v30.xml emd-14566.xml emd-14566.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14566_fsc.xml emd_14566_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14566.png emd_14566.png | 169.6 KB | ||
| Filedesc metadata |  emd-14566.cif.gz emd-14566.cif.gz | 5.9 KB | ||
| Others |  emd_14566_half_map_1.map.gz emd_14566_half_map_1.map.gz emd_14566_half_map_2.map.gz emd_14566_half_map_2.map.gz | 408.5 MB 408.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14566 http://ftp.pdbj.org/pub/emdb/structures/EMD-14566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14566 | HTTPS FTP |
-Validation report
| Summary document |  emd_14566_validation.pdf.gz emd_14566_validation.pdf.gz | 1019.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14566_full_validation.pdf.gz emd_14566_full_validation.pdf.gz | 1019 KB | Display | |
| Data in XML |  emd_14566_validation.xml.gz emd_14566_validation.xml.gz | 26.3 KB | Display | |
| Data in CIF |  emd_14566_validation.cif.gz emd_14566_validation.cif.gz | 35.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14566 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14566 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14566 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14566 | HTTPS FTP |
-Related structure data
| Related structure data |  7z90MC  7ns2C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14566.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14566.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Virion structure | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14566_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14566_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Leishmania RNA virus 1 - 4
| Entire | Name:  Leishmania RNA virus 1 - 4 Leishmania RNA virus 1 - 4 |
|---|---|
| Components |
|
-Supramolecule #1: Leishmania RNA virus 1 - 4
| Supramolecule | Name: Leishmania RNA virus 1 - 4 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 12530 / Sci species name: Leishmania RNA virus 1 - 4 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Leishmania guyanensis (eukaryote) / Strain: MHOM/BR/75/M4147 Leishmania guyanensis (eukaryote) / Strain: MHOM/BR/75/M4147 |
| Molecular weight | Theoretical: 9.72 MDa |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 420.0 Å / T number (triangulation number): 2 |
-Macromolecule #1: Capsid protein,Major capsid protein
| Macromolecule | Name: Capsid protein,Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Leishmania RNA virus 1 - 4 Leishmania RNA virus 1 - 4 |
| Molecular weight | Theoretical: 86.431742 KDa |
| Sequence | String: MADIPNSDKI ACGRKPMFCE IIKLANRKRL IFNTTDERVY DARLNYCSTA DSTVQADCHI YWRLKLRRTD AVFEEYTGQG YSLDTAAYP QQYTDIIRGY YSKHVSSSLA ANTQHFVNVL AMLRHAGACI AHYCMTGKID FDILSKKKHK NKEVVTLSNA D SLSFLPHS ...String: MADIPNSDKI ACGRKPMFCE IIKLANRKRL IFNTTDERVY DARLNYCSTA DSTVQADCHI YWRLKLRRTD AVFEEYTGQG YSLDTAAYP QQYTDIIRGY YSKHVSSSLA ANTQHFVNVL AMLRHAGACI AHYCMTGKID FDILSKKKHK NKEVVTLSNA D SLSFLPHS ALYLPSPLRA SDPEIFNMLY LLGCACDASI AMDNISNTSG AAKYSMPHYN PLQLSHALHV TIFYMLSLMD SC GYGDDAV LALTSGLHSV TTVIAHSDEG GITRDALREL SYTQPYGTMP VPIAGYFQHI NVLFTTQPAW DQFAGIWDYV ILA TAALVH LSDPGMTVND VTYPTTLTTK VATVDGRNSD LAAQMMHSAT RFCDIFVENL STFWGVVANP DGNASQALLH AFNI VACAV EPNRHLEMNV MAPWYWVESS ALFCDYAPFR SPISSAGYGP QCVYGARLVL AATNSLEFTG EAGDYSAYRF EWTTM RHNP LFNILNKRVG DGLANVDFRL RPFNEWLLEG QPSRRSCNSA GHGTPTATCS HKTPNHDTLD EYIWGSTSCD LFHPAE LTS YTAVCVRFRN YLSGADGDVR ILNTPTREVI EGNVVTRCDG IRCLDSNKRI QHVPEVARRY CMMARYLAQA RTFGALT IG DDIIRGFDKV EKIVKMHKSN NRLDQMPLID VTGLCQPMIE TSTVRASTTT RIDPNKLAAA TARVELPLAP RCTSSLIP S SDTVPEPEPQ VGEPGDNGGC APDLGTGGGS GIEGRGSMDI GDPNSVQALA RLQASSVDKL AAALE UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 4542 / Average electron dose: 1.98 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)