[English] 日本語
 Yorodumi
Yorodumi- EMDB-14528: Cryo-EM structure of the whole photosynthetic complex from the gr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the whole photosynthetic complex from the green sulfur bacteria | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | reaction centre / electron transport / energy transfer / green sulfur bacterium / membrane protein / light-harvesting protein complex / PHOTOSYNTHESIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationthylakoid / bacteriochlorophyll binding / iron-sulfur cluster binding / photosynthesis / electron transfer activity / heme binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Chlorobaculum tepidum TLS (bacteria) Chlorobaculum tepidum TLS (bacteria) | |||||||||
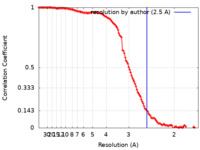
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Xie H / Tsiotis G | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Cryo-EM structure of the whole photosynthetic reaction center apparatus from the green sulfur bacterium . Authors: Hao Xie / Alexandros Lyratzakis / Radhika Khera / Myrto Koutantou / Sonja Welsch / Hartmut Michel / Georgios Tsiotis /   Abstract: Light energy absorption and transfer are very important processes in photosynthesis. In green sulfur bacteria light is absorbed primarily by the chlorosomes and its energy is transferred via the ...Light energy absorption and transfer are very important processes in photosynthesis. In green sulfur bacteria light is absorbed primarily by the chlorosomes and its energy is transferred via the Fenna-Matthews-Olson (FMO) proteins to a homodimeric reaction center (RC). Here, we report the cryogenic electron microscopic structure of the intact FMO-RC apparatus from at 2.5 Å resolution. The FMO-RC apparatus presents an asymmetric architecture and contains two FMO trimers that show different interaction patterns with the RC core. Furthermore, the two permanently bound transmembrane subunits PscC, which donate electrons to the special pair, interact only with the two large PscA subunits. This structure fills an important gap in our understanding of the transfer of energy from antenna to the electron transport chain of this RC and the transfer of electrons from reduced sulfur compounds to the special pair. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14528.map.gz emd_14528.map.gz | 168.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14528-v30.xml emd-14528-v30.xml emd-14528.xml emd-14528.xml | 28.8 KB 28.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14528_fsc.xml emd_14528_fsc.xml | 12.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_14528.png emd_14528.png | 143.3 KB | ||
| Filedesc metadata |  emd-14528.cif.gz emd-14528.cif.gz | 8 KB | ||
| Others |  emd_14528_half_map_1.map.gz emd_14528_half_map_1.map.gz emd_14528_half_map_2.map.gz emd_14528_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14528 http://ftp.pdbj.org/pub/emdb/structures/EMD-14528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14528 | HTTPS FTP |
-Validation report
| Summary document |  emd_14528_validation.pdf.gz emd_14528_validation.pdf.gz | 995.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14528_full_validation.pdf.gz emd_14528_full_validation.pdf.gz | 995.3 KB | Display | |
| Data in XML |  emd_14528_validation.xml.gz emd_14528_validation.xml.gz | 20.3 KB | Display | |
| Data in CIF |  emd_14528_validation.cif.gz emd_14528_validation.cif.gz | 26.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14528 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14528 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14528 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14528 | HTTPS FTP |
-Related structure data
| Related structure data |  7z6qMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14528.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14528.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||
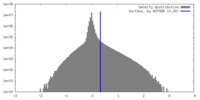
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14528_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
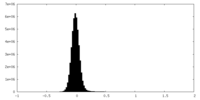
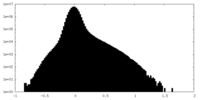
| Density Histograms |
-Half map: #1
| File | emd_14528_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
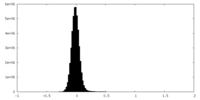
| Density Histograms |
- Sample components
Sample components
+Entire : Photosystem P840 reaction center
+Supramolecule #1: Photosystem P840 reaction center
+Macromolecule #1: Photosystem P840 reaction center, large subunit
+Macromolecule #2: Photosystem P840 reaction center iron-sulfur protein
+Macromolecule #3: Cytochrome c
+Macromolecule #4: P840 reaction center 17 kDa protein
+Macromolecule #5: Bacteriochlorophyll a protein
+Macromolecule #6: Bacteriochlorophyll A isomer
+Macromolecule #7: CHLOROPHYLL A
+Macromolecule #8: BACTERIOCHLOROPHYLL A
+Macromolecule #9: [(2R,3S,4S,5R,6R)-6-[(10E,12E,14E)-2,6,10,14,19,23-hexamethyl-25-...
+Macromolecule #10: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #11: [(2~{R})-2-hexadecanoyloxy-3-[(2~{S},3~{S},4~{R},5~{R},6~{S})-6-(...
+Macromolecule #12: CALCIUM ION
+Macromolecule #13: IRON/SULFUR CLUSTER
+Macromolecule #14: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 4092 pixel / Digitization - Dimensions - Height: 5769 pixel / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.2 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)