+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mammalian Dicer in the dicing state with pre-miR-15a substrate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | endoribonuclease / dsRNA / complex / gene silencing / post-transcriptional / catalytic complex / cytoplasm / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of muscle cell apoptotic process / MicroRNA (miRNA) biogenesis / Small interfering RNA (siRNA) biogenesis / ganglion development / hair follicle cell proliferation / zygote asymmetric cell division / cardiac neural crest cell development involved in outflow tract morphogenesis / regulation of oligodendrocyte differentiation / olfactory bulb interneuron differentiation / regulation of odontogenesis of dentin-containing tooth ...regulation of muscle cell apoptotic process / MicroRNA (miRNA) biogenesis / Small interfering RNA (siRNA) biogenesis / ganglion development / hair follicle cell proliferation / zygote asymmetric cell division / cardiac neural crest cell development involved in outflow tract morphogenesis / regulation of oligodendrocyte differentiation / olfactory bulb interneuron differentiation / regulation of odontogenesis of dentin-containing tooth / trophectodermal cell proliferation / regulation of enamel mineralization / regulation of miRNA metabolic process / peripheral nervous system myelin formation / spermatogonial cell division / regulation of RNA metabolic process / regulation of epithelial cell differentiation / global gene silencing by mRNA cleavage / spinal cord motor neuron differentiation / negative regulation of Schwann cell proliferation / epidermis morphogenesis / reproductive structure development / ribonuclease III / regulation of Notch signaling pathway / myoblast differentiation involved in skeletal muscle regeneration / nerve development / regulation of regulatory T cell differentiation / positive regulation of Schwann cell differentiation / RISC-loading complex / positive regulation of myelination / meiotic spindle organization / intestinal epithelial cell development / RISC complex assembly / regulatory ncRNA-mediated post-transcriptional gene silencing / miRNA processing / ribonuclease III activity / pre-miRNA processing / pericentric heterochromatin formation / siRNA processing / regulation of stem cell differentiation / regulation of viral genome replication / RISC complex / inner ear receptor cell development / mRNA stabilization / cartilage development / embryonic limb morphogenesis / embryonic hindlimb morphogenesis / digestive tract development / positive regulation of miRNA metabolic process / cardiac muscle cell development / miRNA binding / regulation of neuron differentiation / regulation of myelination / negative regulation of glial cell proliferation / hair follicle morphogenesis / stem cell population maintenance / branching morphogenesis of an epithelial tube / hair follicle development / regulation of neurogenesis / RNA processing / postsynaptic density, intracellular component / spindle assembly / spleen development / neuron projection morphogenesis / lung development / post-embryonic development / helicase activity / cerebral cortex development / multicellular organism growth / rRNA processing / regulation of gene expression / regulation of inflammatory response / angiogenesis / defense response to virus / gene expression / cell population proliferation / regulation of cell cycle / positive regulation of gene expression / perinuclear region of cytoplasm / glutamatergic synapse / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / DNA binding / ATP binding / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
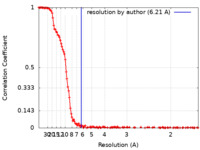
| Method | single particle reconstruction / cryo EM / Resolution: 6.21 Å | |||||||||
 Authors Authors | Zanova M / Zapletal D | |||||||||
| Funding support |  Czech Republic, 2 items Czech Republic, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Structural and functional basis of mammalian microRNA biogenesis by Dicer. Authors: David Zapletal / Eliska Taborska / Josef Pasulka / Radek Malik / Karel Kubicek / Martina Zanova / Christian Much / Marek Sebesta / Valeria Buccheri / Filip Horvat / Irena Jenickova / ...Authors: David Zapletal / Eliska Taborska / Josef Pasulka / Radek Malik / Karel Kubicek / Martina Zanova / Christian Much / Marek Sebesta / Valeria Buccheri / Filip Horvat / Irena Jenickova / Michaela Prochazkova / Jan Prochazka / Matyas Pinkas / Jiri Novacek / Diego F Joseph / Radislav Sedlacek / Carrie Bernecky / Dónal O'Carroll / Richard Stefl / Petr Svoboda /      Abstract: MicroRNA (miRNA) and RNA interference (RNAi) pathways rely on small RNAs produced by Dicer endonucleases. Mammalian Dicer primarily supports the essential gene-regulating miRNA pathway, but how it is ...MicroRNA (miRNA) and RNA interference (RNAi) pathways rely on small RNAs produced by Dicer endonucleases. Mammalian Dicer primarily supports the essential gene-regulating miRNA pathway, but how it is specifically adapted to miRNA biogenesis is unknown. We show that the adaptation entails a unique structural role of Dicer's DExD/H helicase domain. Although mice tolerate loss of its putative ATPase function, the complete absence of the domain is lethal because it assures high-fidelity miRNA biogenesis. Structures of murine Dicer•-miRNA precursor complexes revealed that the DExD/H domain has a helicase-unrelated structural function. It locks Dicer in a closed state, which facilitates miRNA precursor selection. Transition to a cleavage-competent open state is stimulated by Dicer-binding protein TARBP2. Absence of the DExD/H domain or its mutations unlocks the closed state, reduces substrate selectivity, and activates RNAi. Thus, the DExD/H domain structurally contributes to mammalian miRNA biogenesis and underlies mechanistical partitioning of miRNA and RNAi pathways. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14384.map.gz emd_14384.map.gz | 202.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14384-v30.xml emd-14384-v30.xml emd-14384.xml emd-14384.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14384_fsc.xml emd_14384_fsc.xml | 17.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_14384.png emd_14384.png | 55.7 KB | ||
| Filedesc metadata |  emd-14384.cif.gz emd-14384.cif.gz | 7.3 KB | ||
| Others |  emd_14384_half_map_1.map.gz emd_14384_half_map_1.map.gz emd_14384_half_map_2.map.gz emd_14384_half_map_2.map.gz | 199.9 MB 199.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14384 http://ftp.pdbj.org/pub/emdb/structures/EMD-14384 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14384 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14384 | HTTPS FTP |
-Validation report
| Summary document |  emd_14384_validation.pdf.gz emd_14384_validation.pdf.gz | 717.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14384_full_validation.pdf.gz emd_14384_full_validation.pdf.gz | 716.7 KB | Display | |
| Data in XML |  emd_14384_validation.xml.gz emd_14384_validation.xml.gz | 21.2 KB | Display | |
| Data in CIF |  emd_14384_validation.cif.gz emd_14384_validation.cif.gz | 28 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14384 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14384 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14384 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14384 | HTTPS FTP |
-Related structure data
| Related structure data |  7yynMC  7yymC  7yz4C  7zpiC  7zpjC  7zpkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14384.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14384.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14384_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14384_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dicing state of mouse oocyte dicer with pre-miR-15a
| Entire | Name: Dicing state of mouse oocyte dicer with pre-miR-15a |
|---|---|
| Components |
|
-Supramolecule #1: Dicing state of mouse oocyte dicer with pre-miR-15a
| Supramolecule | Name: Dicing state of mouse oocyte dicer with pre-miR-15a / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: 59-nt precursor of miR-15a
| Supramolecule | Name: 59-nt precursor of miR-15a / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Isoform 2 of Endoribonuclease Dicer
| Supramolecule | Name: Isoform 2 of Endoribonuclease Dicer / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: 59-nt precursor of miR-15a
| Macromolecule | Name: 59-nt precursor of miR-15a / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.017279 KDa |
| Sequence | String: UAGCAGCACA UAAUGGUUUG UGGAUGUUGA AAAGGUGCAG GCCAUACUGU GCUGCCUCA GENBANK: GENBANK: AC154660.2 |
-Macromolecule #2: Isoform 2 of Endoribonuclease Dicer
| Macromolecule | Name: Isoform 2 of Endoribonuclease Dicer / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: ribonuclease III |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 200.040938 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MVWSHPQFEK GGGSGGGSGG SAWSHPQFEK GDYPYDVPDY AGTENLYFQG LVDSRDTEVY TSQPCEIVVD CGPFTDRSGL YERLLMELE AALDFINDCN VAVHSKERDS TLISKQILSD CRAVLVVLGP WCADKVAGMM VRELQKYIKH EQEELHRKFL L FTDTLLRK ...String: MVWSHPQFEK GGGSGGGSGG SAWSHPQFEK GDYPYDVPDY AGTENLYFQG LVDSRDTEVY TSQPCEIVVD CGPFTDRSGL YERLLMELE AALDFINDCN VAVHSKERDS TLISKQILSD CRAVLVVLGP WCADKVAGMM VRELQKYIKH EQEELHRKFL L FTDTLLRK IHALCEEYFS PASLDLKYVT PKVMKLLEIL RKYKPYERQQ FESVEWYNNR NQDNYVSWSD SEDDDDDEEI EE KEKPETN FPSPFTNILC GIIFVERRYT AVVLNRLIKE AGKQDPELAY ISSNFITGHG IGKNQPRSKQ MEAEFRKQEE VLR KFRAHE TNLLIATSVV EEGVDIPKCN LVVRFDLPTE YRSYVQSKGR ARAPISNYVM LADTDKIKSF EEDLKTYKAI EKIL RNKCS KSADGAEADV HAGVDDEDAF PPYVLRPDDG GPRVTINTAI GHINRYCARL PSDPFTHLAP KCRTRELPDG TFYST LYLP INSPLRASIV GPPMDSVRLA ERVVALICCE KLHKIGELDE HLMPVGKETV KYEEELDLHD EEETSVPGRP GSTKRR QCY PKAIPECLRE SYPKPDQPCY LYVIGMVLTT PLPDELNFRR RKLYPPEDTT RCFGILTAKP IPQIPHFPVY TRSGEVT IS IELKKSGFTL SQQMLELITR LHQYIFSHIL RLEKPALEFK PTGAESAYCV LPLNVVNDSG TLDIDFKFME DIEKSEAR I GIPSTKYSKE TPFVFKLEDY QDAVIIPRYR NFDQPHRFYV ADVYTDLTPL SKFPSPEYET FAEYYKTKYN LDLTNLNQP LLDVDHTSSR LNLLTPRHLN QKGKALPLSS AEKRKAKWES LQNKQILVPE LCAIHPIPAS LWRKAVCLPS ILYRLHCLLT AEELRAQTA SDAGVGVRSL PVDFRYPNLD FGWKKSIDSK SFISSCNSSL AESDNYCKHS TTVVPEHAAH QGATRPSLEN H DQMSVNCK RLPAESPAKL QSEVSTDLTA INGLSYNKNL ANGSYDLVNR DFCQGNQLNY FKQEIPVQPT TSYPIQNLYN YE NQPKPSN ECPLLSNTYL DGNANTSTSD GSPAVSTMPA MMNAVKALKD RMDSEQSPSV GYSSRTLGPN PGLILQALTL SNA SDGFNL ERLEMLGDSF LKHAITTYLF CTYPDAHEGR LSYMRSKKVS NCNLYRLGKK KGLPSRMVVS IFDPPVNWLP PGYV VNQDK SNSEKWEKDE MTKDCLLANG KLGEACEEEE DLTWRAPKEE AEDEDDFLEY DQEHIQFIDS MLMGSGAFVR KISLS PFSA SDSAYEWKMP KKASLGSMPF ASGLEDFDYS SWDAMCYLDP SKAVEEDDFV VGFWNPSEEN CGVDTGKQSI SYDLHT EQC IADKSIADCV AALLGCYLTS CGERAAQLFL CSLGLKVLPV IKRTSREKAL DPAQENGSSQ QKSLSGSCAS PVGPRSS AG KDLEYGCLKI PPRCMFDHPD AEKTLNHLIS GFETFEKKIN YRFKNKAYLL QAFTHASYHY NTITDCYQRL EFLGDAIL D YLITKHLYED PRQHSPGVLT DLRSALVNNT IFASLAVKYD YHKYFKAVSP ELFHVIDDFV KFQLEKNEMQ GMDSELRRS EEDEEKEEDI EVPKAMGDIF ASLAGAIYMD SGMSLEVVWQ VYYPMMQPLI EKFSANVPRS PVRELLEMEP ETAKFSPAER TYDGKVRVT VEVVGKGKFK GVGRSYRIAK SAAARRALRS LKANQPQVPN SGRGENLYFQ GASDYKDHDG DYKDHDGSHH H HHHHH UniProtKB: Endoribonuclease Dicer |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.20 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: The buffer was always fresh in RNAse-free manner | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: Described in STAR methods. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 9956 / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Initial model for mouse oocyte Dicer was calculated using AlphaFold 2.0. Initial rigid body fitting was done using Chimera. Further refinement was done using ISOLDE with distance restrains |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 451.4 / Target criteria: Correlation coefficient |
| Output model |  PDB-7yyn: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)