+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1435 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Functional architecture of RNA polymerase I. | |||||||||
 Map data Map data | This is the final reconstruction of RNA polymerase I in ccp4 format. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | RNA polymerase I complex Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 11.9 Å | |||||||||
 Authors Authors | Kuhn C-D / Geiger SR / Baumli S / Gartmann M / Gerber J / Jennebach S / Mielke T / Tschochner H / Beckmann R / Cramer P | |||||||||
 Citation Citation |  Journal: Cell / Year: 2007 Journal: Cell / Year: 2007Title: Functional architecture of RNA polymerase I. Authors: Claus-D Kuhn / Sebastian R Geiger / Sonja Baumli / Marco Gartmann / Jochen Gerber / Stefan Jennebach / Thorsten Mielke / Herbert Tschochner / Roland Beckmann / Patrick Cramer /  Abstract: Synthesis of ribosomal RNA (rRNA) by RNA polymerase (Pol) I is the first step in ribosome biogenesis and a regulatory switch in eukaryotic cell growth. Here we report the 12 A cryo-electron ...Synthesis of ribosomal RNA (rRNA) by RNA polymerase (Pol) I is the first step in ribosome biogenesis and a regulatory switch in eukaryotic cell growth. Here we report the 12 A cryo-electron microscopic structure for the complete 14-subunit yeast Pol I, a homology model for the core enzyme, and the crystal structure of the subcomplex A14/43. In the resulting hybrid structure of Pol I, A14/43, the clamp, and the dock domain contribute to a unique surface interacting with promoter-specific initiation factors. The Pol I-specific subunits A49 and A34.5 form a heterodimer near the enzyme funnel that acts as a built-in elongation factor and is related to the Pol II-associated factor TFIIF. In contrast to Pol II, Pol I has a strong intrinsic 3'-RNA cleavage activity, which requires the C-terminal domain of subunit A12.2 and, apparently, enables ribosomal RNA proofreading and 3'-end trimming. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1435.map.gz emd_1435.map.gz | 710.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1435-v30.xml emd-1435-v30.xml emd-1435.xml emd-1435.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| Images |  1435.gif 1435.gif | 20.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1435 http://ftp.pdbj.org/pub/emdb/structures/EMD-1435 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1435 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1435 | HTTPS FTP |
-Validation report
| Summary document |  emd_1435_validation.pdf.gz emd_1435_validation.pdf.gz | 212.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1435_full_validation.pdf.gz emd_1435_full_validation.pdf.gz | 211.7 KB | Display | |
| Data in XML |  emd_1435_validation.xml.gz emd_1435_validation.xml.gz | 5.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1435 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1435 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1435 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1435 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1435.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1435.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the final reconstruction of RNA polymerase I in ccp4 format. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
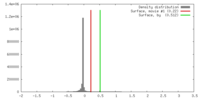
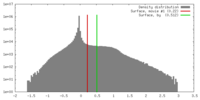
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : S. cerevisiae RNA polymerase I
| Entire | Name: S. cerevisiae RNA polymerase I |
|---|---|
| Components |
|
-Supramolecule #1000: S. cerevisiae RNA polymerase I
| Supramolecule | Name: S. cerevisiae RNA polymerase I / type: sample / ID: 1000 / Oligomeric state: monomeric / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 600 KDa / Theoretical: 600 KDa |
-Macromolecule #1: RNA polymerase I
| Macromolecule | Name: RNA polymerase I / type: protein_or_peptide / ID: 1 / Name.synonym: DNA-dependant RNA polymerase I / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 600 KDa / Theoretical: 600 KDa |
| Recombinant expression | Organism:  |
| Sequence | GO: RNA polymerase I complex |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.8 Details: 60mM ammonium sulfate, 5mM HEPES pH 7.8, 1mM magnesium chloride, 0.1mM zinc chloride |
| Staining | Type: NEGATIVE / Details: Vitrification |
| Grid | Details: Carbon holey grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: OTHER / Details: Vitrification instrument: Vitrobot |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Temperature | Average: 100 K |
| Date | Mar 20, 2006 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: PRIMESCAN / Average electron dose: 20 e/Å2 / Details: Heidelberg Drum Scanner / Bits/pixel: 16 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal magnification: 39000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Quantifoil |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 11.9 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER / Number images used: 46056 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Details | Protocol: rigid body. 1WCM (lacking Rpb4/7 and the foot domain) was fitted by manual docking using program O. |
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)