+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
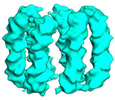
| Title | Six DNA Helix Bundle nanopore - State 5 | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | DNA origami / nanopore. / DNA | |||||||||||||||
| Biological species | DNA molecule (others) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.0 Å | |||||||||||||||
 Authors Authors | Javed A / Ahmad K | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations. Authors: Katya Ahmad / Abid Javed / Conor Lanphere / Peter V Coveney / Elena V Orlova / Stefan Howorka /   Abstract: DNA can be folded into rationally designed, unique, and functional materials. To fully realise the potential of these DNA materials, a fundamental understanding of their structure and dynamics is ...DNA can be folded into rationally designed, unique, and functional materials. To fully realise the potential of these DNA materials, a fundamental understanding of their structure and dynamics is necessary, both in simple solvents as well as more complex and diverse anisotropic environments. Here we analyse an archetypal six-duplex DNA nanoarchitecture with single-particle cryo-electron microscopy and molecular dynamics simulations in solvents of tunable ionic strength and within the anisotropic environment of biological membranes. Outside lipid bilayers, the six-duplex bundle lacks the designed symmetrical barrel-type architecture. Rather, duplexes are arranged in non-hexagonal fashion and are disorted to form a wider, less elongated structure. Insertion into lipid membranes, however, restores the anticipated barrel shape due to lateral duplex compression by the bilayer. The salt concentration has a drastic impact on the stability of the inserted barrel-shaped DNA nanopore given the tunable electrostatic repulsion between the negatively charged duplexes. By synergistically combining experiments and simulations, we increase fundamental understanding into the environment-dependent structural dynamics of a widely used nanoarchitecture. This insight will pave the way for future engineering and biosensing applications. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14346.map.gz emd_14346.map.gz | 51.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14346-v30.xml emd-14346-v30.xml emd-14346.xml emd-14346.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14346.png emd_14346.png | 26.1 KB | ||
| Filedesc metadata |  emd-14346.cif.gz emd-14346.cif.gz | 5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14346 http://ftp.pdbj.org/pub/emdb/structures/EMD-14346 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14346 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14346 | HTTPS FTP |
-Related structure data
| Related structure data |  7ywoMC  7ywhC  7ywiC  7ywlC  7ywnC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14346.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14346.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
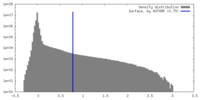
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Six DNA helix bundle DNA nanopore
| Entire | Name: Six DNA helix bundle DNA nanopore |
|---|---|
| Components |
|
-Supramolecule #1: Six DNA helix bundle DNA nanopore
| Supramolecule | Name: Six DNA helix bundle DNA nanopore / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Macromolecule #1: DNA (50-MER)
| Macromolecule | Name: DNA (50-MER) / type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 15.334821 KDa |
| Sequence | String: (DA)(DG)(DC)(DG)(DA)(DA)(DC)(DG)(DT)(DG) (DG)(DA)(DT)(DT)(DT)(DT)(DG)(DT)(DC)(DC) (DG)(DA)(DC)(DA)(DT)(DC)(DG)(DG)(DC) (DA)(DA)(DG)(DC)(DT)(DC)(DC)(DC)(DT)(DT) (DT) (DT)(DT)(DC)(DG)(DA)(DC)(DT)(DA) (DT)(DT) |
-Macromolecule #2: DNA (50-MER)
| Macromolecule | Name: DNA (50-MER) / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 15.413827 KDa |
| Sequence | String: (DC)(DC)(DG)(DA)(DT)(DG)(DT)(DC)(DG)(DG) (DA)(DC)(DT)(DT)(DT)(DT)(DA)(DC)(DA)(DC) (DG)(DA)(DT)(DC)(DT)(DT)(DC)(DG)(DC) (DC)(DT)(DG)(DC)(DT)(DG)(DG)(DG)(DT)(DT) (DT) (DT)(DG)(DG)(DG)(DA)(DG)(DC)(DT) (DT)(DG) |
-Macromolecule #3: DNA (50-MER)
| Macromolecule | Name: DNA (50-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 15.285784 KDa |
| Sequence | String: (DC)(DG)(DA)(DA)(DG)(DA)(DT)(DC)(DG)(DT) (DG)(DT)(DT)(DT)(DT)(DT)(DC)(DC)(DA)(DC) (DA)(DG)(DT)(DT)(DG)(DA)(DT)(DT)(DG) (DC)(DC)(DC)(DT)(DT)(DC)(DA)(DC)(DT)(DT) (DT) (DT)(DC)(DC)(DC)(DA)(DG)(DC)(DA) (DG)(DG) |
-Macromolecule #4: DNA (50-MER)
| Macromolecule | Name: DNA (50-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 15.483923 KDa |
| Sequence | String: (DA)(DA)(DT)(DC)(DA)(DA)(DC)(DT)(DG)(DT) (DG)(DG)(DT)(DT)(DT)(DT)(DT)(DC)(DT)(DC) (DA)(DC)(DT)(DG)(DG)(DT)(DG)(DA)(DT) (DT)(DA)(DG)(DA)(DA)(DT)(DG)(DC)(DT)(DT) (DT) (DT)(DG)(DT)(DG)(DA)(DA)(DG)(DG) (DG)(DC) |
-Macromolecule #5: DNA (50-MER)
| Macromolecule | Name: DNA (50-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 15.403877 KDa |
| Sequence | String: (DT)(DC)(DA)(DC)(DC)(DA)(DG)(DT)(DG)(DA) (DG)(DA)(DT)(DT)(DT)(DT)(DT)(DG)(DT)(DC) (DG)(DT)(DA)(DC)(DC)(DA)(DG)(DG)(DT) (DG)(DC)(DA)(DT)(DG)(DG)(DA)(DT)(DT)(DT) (DT) (DT)(DG)(DC)(DA)(DT)(DT)(DC)(DT) (DA)(DA) |
-Macromolecule #6: DNA (50-MER)
| Macromolecule | Name: DNA (50-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 15.268795 KDa |
| Sequence | String: (DC)(DC)(DT)(DG)(DG)(DT)(DA)(DC)(DG)(DA) (DC)(DA)(DT)(DT)(DT)(DT)(DT)(DC)(DC)(DA) (DC)(DG)(DT)(DT)(DC)(DG)(DC)(DT)(DA) (DA)(DT)(DA)(DG)(DT)(DC)(DG)(DA)(DT)(DT) (DT) (DT)(DA)(DT)(DC)(DC)(DA)(DT)(DG) (DC)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Concentration: 12.0 mM / Component - Formula: MgCl2 / Component - Name: Magnesium Chloride |
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average electron dose: 1.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 47000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Cross-correlation |
|---|---|
| Output model |  PDB-7ywo: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)