+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Bacillus megaterium gas vesicles | |||||||||
 Map data Map data | Sharpened cryo-EM density - cut to 128px box size around the part optimized in local refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | gas vesicle / buoyancy / helical / microbial motility / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Huber ST / Evers W / Jakobi AJ | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Cryo-EM structure of gas vesicles for buoyancy-controlled motility. Authors: Stefan T Huber / Dion Terwiel / Wiel H Evers / David Maresca / Arjen J Jakobi /  Abstract: Gas vesicles are gas-filled nanocompartments that allow a diverse group of bacteria and archaea to control their buoyancy. The molecular basis of their properties and assembly remains unclear. Here, ...Gas vesicles are gas-filled nanocompartments that allow a diverse group of bacteria and archaea to control their buoyancy. The molecular basis of their properties and assembly remains unclear. Here, we report the 3.2 Å cryo-EM structure of the gas vesicle shell made from the structural protein GvpA that self-assembles into hollow helical cylinders closed off by cone-shaped tips. Two helical half shells connect through a characteristic arrangement of GvpA monomers, suggesting a mechanism of gas vesicle biogenesis. The fold of GvpA features a corrugated wall structure typical for force-bearing thin-walled cylinders. Small pores enable gas molecules to diffuse across the shell, while the exceptionally hydrophobic interior surface effectively repels water. Comparative structural analysis confirms the evolutionary conservation of gas vesicle assemblies and demonstrates molecular features of shell reinforcement by GvpC. Our findings will further research into gas vesicle biology and facilitate molecular engineering of gas vesicles for ultrasound imaging. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14238.map.gz emd_14238.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14238-v30.xml emd-14238-v30.xml emd-14238.xml emd-14238.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14238_fsc.xml emd_14238_fsc.xml | 17.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_14238.png emd_14238.png | 245.5 KB | ||
| Masks |  emd_14238_msk_1.map emd_14238_msk_1.map | 8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14238.cif.gz emd-14238.cif.gz | 6.3 KB | ||
| Others |  emd_14238_additional_1.map.gz emd_14238_additional_1.map.gz emd_14238_additional_2.map.gz emd_14238_additional_2.map.gz emd_14238_half_map_1.map.gz emd_14238_half_map_1.map.gz emd_14238_half_map_2.map.gz emd_14238_half_map_2.map.gz | 115.6 MB 7.5 MB 7.5 MB 7.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14238 http://ftp.pdbj.org/pub/emdb/structures/EMD-14238 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14238 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14238 | HTTPS FTP |
-Validation report
| Summary document |  emd_14238_validation.pdf.gz emd_14238_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14238_full_validation.pdf.gz emd_14238_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_14238_validation.xml.gz emd_14238_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_14238_validation.cif.gz emd_14238_validation.cif.gz | 22.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14238 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14238 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14238 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14238 | HTTPS FTP |
-Related structure data
| Related structure data |  7r1cMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14238.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14238.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened cryo-EM density - cut to 128px box size around the part optimized in local refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||||||||||||||||||
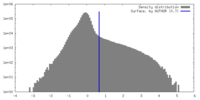
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14238_msk_1.map emd_14238_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
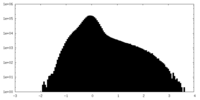
| Density Histograms |
-Additional map: Symmetrized cryo-EM density before local refinement / helical...
| File | emd_14238_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Symmetrized cryo-EM density before local refinement / helical twist is -3.874 degrees / helical rise is 0.525 Angstroms. | ||||||||||||
| Projections & Slices |
| ||||||||||||
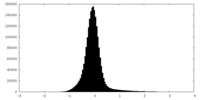
| Density Histograms |
-Additional map: Unsharpened cryo-EM density - cut to 128px box...
| File | emd_14238_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened cryo-EM density - cut to 128px box size around the part optimized in local refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
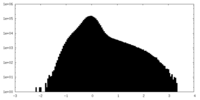
| Density Histograms |
-Half map: Half-map B - cut to 128px box size...
| File | emd_14238_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B - cut to 128px box size around the part optimized in local refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map A - cut to 128px box size...
| File | emd_14238_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A - cut to 128px box size around the part optimized in local refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Helical assembly of GvpB monomers forming the gas vesicle wall
| Entire | Name: Helical assembly of GvpB monomers forming the gas vesicle wall |
|---|---|
| Components |
|
-Supramolecule #1: Helical assembly of GvpB monomers forming the gas vesicle wall
| Supramolecule | Name: Helical assembly of GvpB monomers forming the gas vesicle wall type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) |
| Molecular weight | Theoretical: 9.99 MDa |
-Macromolecule #1: Gas vesicle structural protein
| Macromolecule | Name: Gas vesicle structural protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria) Priestia megaterium NBRC 15308 = ATCC 14581 (bacteria)Strain: ATCC 14581 / DSM 32 / JCM 2506 / NBRC 15308 / NCIMB 9376 / NCTC 10342 / NRRL B-14308 / VKM B-512 |
| Molecular weight | Theoretical: 9.62685 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSIQKSTNSS SLAEVIDRIL DKGIVIDAFA RVSVVGIEIL TIEARVVIAS VDTWLRYAEA VGLLRDDVEE NGLPERSNSS EGQPRFSI UniProtKB: Gas vesicle protein A |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 0.45 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: LEICA PLUNGER / Details: Blot times between 5 and 11 seconds.. | |||||||||
| Details | Concentration measured by OD(500)=3.12 against a sonicated blank. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 4351 / Average exposure time: 2.4 sec. / Average electron dose: 30.0 e/Å2 Details: One shot per hole 1.37 A/pix 60 fractions over 30 e-/A2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.25 µm / Nominal defocus min: 0.25 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7r1c: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)