+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Abortive infection DNA polymerase AbiK from Lactococcus lactis | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA polymerase / protein-primed DNA synthesis / template-independent DNA synthesis / abortive infection / ANTIVIRAL PROTEIN | |||||||||
| Function / homology | Reverse transcriptase domain / Reverse transcriptase (RNA-dependent DNA polymerase) / Reverse transcriptase (RT) catalytic domain profile. / DNA/RNA polymerase superfamily / metal ion binding / AbiK Function and homology information Function and homology information | |||||||||
| Biological species |  Lactococcus lactis (lactic acid bacteria) / Lactococcus lactis (lactic acid bacteria) /  | |||||||||
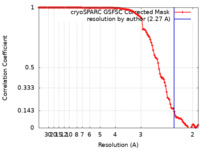
| Method | single particle reconstruction / cryo EM / Resolution: 2.27 Å | |||||||||
 Authors Authors | Figiel M / Nowotny M / Gapinska M / Czarnocki-Cieciura M / Zajko W | |||||||||
| Funding support |  Poland, European Union, 2 items Poland, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases. Authors: Małgorzata Figiel / Marta Gapińska / Mariusz Czarnocki-Cieciura / Weronika Zajko / Małgorzata Sroka / Krzysztof Skowronek / Marcin Nowotny Abstract: Abortive infection (Abi) is a bacterial antiphage defense strategy involving suicide of the infected cell. Some Abi pathways involve polymerases that are related to reverse transcriptases. They are ...Abortive infection (Abi) is a bacterial antiphage defense strategy involving suicide of the infected cell. Some Abi pathways involve polymerases that are related to reverse transcriptases. They are unique in the way they combine the ability to synthesize DNA in a template-independent manner with protein priming. Here, we report crystal and cryo-electron microscopy structures of two Abi polymerases: AbiK and Abi-P2. Both proteins adopt a bilobal structure with an RT-like domain that comprises palm and fingers subdomains and a unique helical domain. AbiK and Abi-P2 adopt a hexameric and trimeric configuration, respectively, which is unprecedented for reverse transcriptases. Biochemical experiments showed that the formation of these oligomers is required for the DNA polymerization activity. The structure of the AbiK-DNA covalent adduct visualized interactions between the 3' end of DNA and the active site and covalent attachment of the 5' end of DNA to a tyrosine residue used for protein priming. Our data reveal a structural basis of the mechanism of highly unusual template-independent protein-priming polymerases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14220.map.gz emd_14220.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14220-v30.xml emd-14220-v30.xml emd-14220.xml emd-14220.xml | 22.4 KB 22.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14220_fsc.xml emd_14220_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14220.png emd_14220.png | 167.4 KB | ||
| Masks |  emd_14220_msk_1.map emd_14220_msk_1.map | 149.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14220.cif.gz emd-14220.cif.gz | 7.4 KB | ||
| Others |  emd_14220_half_map_1.map.gz emd_14220_half_map_1.map.gz emd_14220_half_map_2.map.gz emd_14220_half_map_2.map.gz | 139.1 MB 139.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14220 http://ftp.pdbj.org/pub/emdb/structures/EMD-14220 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14220 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14220 | HTTPS FTP |
-Related structure data
| Related structure data |  7r06MC  7r07C  7r08C  7z0zC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14220.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14220.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
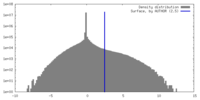
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14220_msk_1.map emd_14220_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
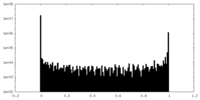
| Density Histograms |
-Half map: #2
| File | emd_14220_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14220_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homohexamer of AbiK with single-stranded DNA covalently attached ...
| Entire | Name: Homohexamer of AbiK with single-stranded DNA covalently attached to each of the subunits |
|---|---|
| Components |
|
-Supramolecule #1: Homohexamer of AbiK with single-stranded DNA covalently attached ...
| Supramolecule | Name: Homohexamer of AbiK with single-stranded DNA covalently attached to each of the subunits type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 420 KDa |
-Supramolecule #2: Abortive infection protein AbiK
| Supramolecule | Name: Abortive infection protein AbiK / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Lactococcus lactis (lactic acid bacteria) Lactococcus lactis (lactic acid bacteria) |
-Supramolecule #3: DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3')
| Supramolecule | Name: DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3') / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: AbiK
| Macromolecule | Name: AbiK / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lactococcus lactis (lactic acid bacteria) Lactococcus lactis (lactic acid bacteria) |
| Molecular weight | Theoretical: 71.713086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSMKKEFTEL YDFIFDPIFL VRYGYYDRSI KNKKMNTAKV ELDNE(PTR)GKSD SFYFKVFNME SFADYLRSHD LKTHFN GKK PLSTDPVYFN IPKNIEARRQ YKMPNLYSYM ALNYYICDNK KEFIEVFIDN KFSTSKFFNQ LNFDYPKTQE ITQTLLY GG IKKLHLDLSN ...String: GSMKKEFTEL YDFIFDPIFL VRYGYYDRSI KNKKMNTAKV ELDNE(PTR)GKSD SFYFKVFNME SFADYLRSHD LKTHFN GKK PLSTDPVYFN IPKNIEARRQ YKMPNLYSYM ALNYYICDNK KEFIEVFIDN KFSTSKFFNQ LNFDYPKTQE ITQTLLY GG IKKLHLDLSN FYHTLYTHSI PWMIDGKSAS KQNRKKGFSN TLDTLITACQ YDETHGIPTG NLLSRIITEL YMCHFDKQ M EYKKFVYSRY VDDFIFPFTF ENEKQEFLNE FNLICRENNL IINDNKTKVD NFPFVDKSSK SDIFSFFENI TSTNSNDKW IKEISNFIDY CVNEEHLGNK GAIKCIFPVI TNTLKQKKVD TKNIDNIFSK RNMVTNFNVF EKILDLSLKD SRLTNKFLTF FENINEFGF SSLSASNIVK KYFSNNSKGL KEKIDHYRKN NFNQELYQIL LYMVVFEIDD LLNQEELLNL IDLNIDDYSL I LGTILYLK NSSYKLEKLL KKIDQLFINT HANYDVKTSR MAEKLWLFRY FFYFLNCKNI FSQKEINSYC QSQNYNSGQN GY QTELNWN YIKGQGKDLR ANNFFNELIV KEVWLISCGE NEDFKYLN UniProtKB: AbiK |
-Macromolecule #2: DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3')
| Macromolecule | Name: DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3') / type: dna / ID: 2 / Number of copies: 6 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 3.425224 KDa |
| Sequence | String: (DC)(DC)(DC)(DC)(DC)(DC)(DC)(DC)(DC)(DC) (DC)(DC) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 150000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)