[English] 日本語
 Yorodumi
Yorodumi- EMDB-14216: Cryo-electron tomography of in vitro assembled Bruno oskar 3'UTR ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-14216 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomography of in vitro assembled Bruno oskar 3'UTR RNA condensates aged for 30 minutes followed by addition of more oskar 3'UTR RNA | |||||||||
 Map data Map data | tomogram of in vitro assembled Bruno oskar 3'UTR RNA condensates that were aged for 30 minutes followed by addition of more oskar 3'UTR RNA. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Bose M / Mahamid J | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Liquid-to-solid phase transition of oskar ribonucleoprotein granules is essential for their function in Drosophila embryonic development. Authors: Mainak Bose / Marko Lampe / Julia Mahamid / Anne Ephrussi /  Abstract: Asymmetric localization of oskar ribonucleoprotein (RNP) granules to the oocyte posterior is crucial for abdominal patterning and germline formation in the Drosophila embryo. We show that oskar RNP ...Asymmetric localization of oskar ribonucleoprotein (RNP) granules to the oocyte posterior is crucial for abdominal patterning and germline formation in the Drosophila embryo. We show that oskar RNP granules in the oocyte are condensates with solid-like physical properties. Using purified oskar RNA and scaffold proteins Bruno and Hrp48, we confirm in vitro that oskar granules undergo a liquid-to-solid phase transition. Whereas the liquid phase allows RNA incorporation, the solid phase precludes incorporation of additional RNA while allowing RNA-dependent partitioning of client proteins. Genetic modification of scaffold granule proteins or tethering the intrinsically disordered region of human fused in sarcoma (FUS) to oskar mRNA allowed modulation of granule material properties in vivo. The resulting liquid-like properties impaired oskar localization and translation with severe consequences on embryonic development. Our study reflects how physiological phase transitions shape RNA-protein condensates to regulate the localization and expression of a maternal RNA that instructs germline formation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14216.map.gz emd_14216.map.gz | 2.3 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14216-v30.xml emd-14216-v30.xml emd-14216.xml emd-14216.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14216.png emd_14216.png | 213.2 KB | ||
| Others |  emd_14216_additional_1.map.gz emd_14216_additional_1.map.gz | 2.3 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14216 http://ftp.pdbj.org/pub/emdb/structures/EMD-14216 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14216 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14216 | HTTPS FTP |
-Validation report
| Summary document |  emd_14216_validation.pdf.gz emd_14216_validation.pdf.gz | 343.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14216_full_validation.pdf.gz emd_14216_full_validation.pdf.gz | 343.4 KB | Display | |
| Data in XML |  emd_14216_validation.xml.gz emd_14216_validation.xml.gz | 4.8 KB | Display | |
| Data in CIF |  emd_14216_validation.cif.gz emd_14216_validation.cif.gz | 5.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14216 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14216 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14216 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14216 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14216.map.gz / Format: CCP4 / Size: 2.5 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14216.map.gz / Format: CCP4 / Size: 2.5 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | tomogram of in vitro assembled Bruno oskar 3'UTR RNA condensates that were aged for 30 minutes followed by addition of more oskar 3'UTR RNA. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 8.51 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
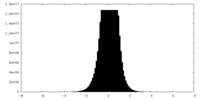
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: tomogram of in vitro assembled Bruno oskar 3'UTR...
| File | emd_14216_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | tomogram of in vitro assembled Bruno oskar 3'UTR RNA condensates that were aged for 30 minutes followed by addition of more oskar 3'UTR RNA. Tomogram was gaussian filtered with a kernel size 3. | ||||||||||||
| Projections & Slices |
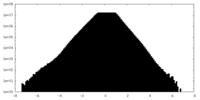
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Condensates assembled using 10 micro molar Bruno-EGFP with 100 na...
| Entire | Name: Condensates assembled using 10 micro molar Bruno-EGFP with 100 nano molar oskar 3'UTR RNA, aged for 30min followed by addition of 400 nano molar oskar 3'UTR RNA |
|---|---|
| Components |
|
-Supramolecule #1: Condensates assembled using 10 micro molar Bruno-EGFP with 100 na...
| Supramolecule | Name: Condensates assembled using 10 micro molar Bruno-EGFP with 100 nano molar oskar 3'UTR RNA, aged for 30min followed by addition of 400 nano molar oskar 3'UTR RNA type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Spodoptera (butterflies/moths) Spodoptera (butterflies/moths) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
| Details | Condensates assembled using 10 micro molar Bruno-EGFP with 100 nano molar oskar 3'UTR RNA, aged for 30min followed by addition of 400 nano molar oskar 3'UTR RNA |
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 2.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 3.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Software - Name:  IMOD / Number images used: 61 IMOD / Number images used: 61 |
|---|
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)