+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
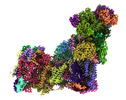
| タイトル | Bovine complex I in lipid nanodisc, Deactive-ligand (composite) | |||||||||
 マップデータ マップデータ | Composite of 3 globally sharpened focus-refined maps generated using Phenix Combine Focused Maps | |||||||||
 試料 試料 |
| |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Complex I biogenesis / Mitochondrial protein import / RHOG GTPase cycle / ubiquinone biosynthetic process / Respiratory electron transport / cellular response to oxygen levels / mitochondrial large ribosomal subunit binding / gliogenesis / neural precursor cell proliferation / [2Fe-2S] cluster assembly ...Complex I biogenesis / Mitochondrial protein import / RHOG GTPase cycle / ubiquinone biosynthetic process / Respiratory electron transport / cellular response to oxygen levels / mitochondrial large ribosomal subunit binding / gliogenesis / neural precursor cell proliferation / [2Fe-2S] cluster assembly / oxygen sensor activity / Neutrophil degranulation / NADH dehydrogenase activity / Mitochondrial protein degradation / acyl binding / mitochondrial ATP synthesis coupled electron transport / ubiquinone binding / acyl carrier activity / electron transport coupled proton transport / NADH:ubiquinone reductase (H+-translocating) / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / respiratory chain complex I / response to cAMP / NADH dehydrogenase (ubiquinone) activity / quinone binding / ATP synthesis coupled electron transport / neurogenesis / reactive oxygen species metabolic process / aerobic respiration / fatty acid binding / respiratory electron transport chain / electron transport chain / circadian rhythm / mitochondrial membrane / brain development / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / NAD binding / fatty acid biosynthetic process / FMN binding / 4 iron, 4 sulfur cluster binding / oxidoreductase activity / mitochondrial inner membrane / mitochondrial matrix / negative regulation of DNA-templated transcription / apoptotic process / protein-containing complex binding / mitochondrion / nucleoplasm / metal ion binding / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |   | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.3 Å | |||||||||
 データ登録者 データ登録者 | Chung I / Bridges HR / Hirst J | |||||||||
| 資金援助 |  英国, 1件 英国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2022 ジャーナル: Nat Commun / 年: 2022タイトル: Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy. 著者: Injae Chung / John J Wright / Hannah R Bridges / Bozhidar S Ivanov / Olivier Biner / Caroline S Pereira / Guilherme M Arantes / Judy Hirst /    要旨: Mitochondrial complex I is a central metabolic enzyme that uses the reducing potential of NADH to reduce ubiquinone-10 (Q) and drive four protons across the inner mitochondrial membrane, powering ...Mitochondrial complex I is a central metabolic enzyme that uses the reducing potential of NADH to reduce ubiquinone-10 (Q) and drive four protons across the inner mitochondrial membrane, powering oxidative phosphorylation. Although many complex I structures are now available, the mechanisms of Q reduction and energy transduction remain controversial. Here, we reconstitute mammalian complex I into phospholipid nanodiscs with exogenous Q. Using cryo-EM, we reveal a Q molecule occupying the full length of the Q-binding site in the 'active' (ready-to-go) resting state together with a matching substrate-free structure, and apply molecular dynamics simulations to propose how the charge states of key residues influence the Q binding pose. By comparing ligand-bound and ligand-free forms of the 'deactive' resting state (that require reactivating to catalyse), we begin to define how substrate binding restructures the deactive Q-binding site, providing insights into its physiological and mechanistic relevance. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_14134.map.gz emd_14134.map.gz | 929.3 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-14134-v30.xml emd-14134-v30.xml emd-14134.xml emd-14134.xml | 68 KB 68 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_14134.png emd_14134.png | 137.4 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14134 http://ftp.pdbj.org/pub/emdb/structures/EMD-14134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14134 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_14134_validation.pdf.gz emd_14134_validation.pdf.gz | 546 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_14134_full_validation.pdf.gz emd_14134_full_validation.pdf.gz | 545.6 KB | 表示 | |
| XML形式データ |  emd_14134_validation.xml.gz emd_14134_validation.xml.gz | 9 KB | 表示 | |
| CIF形式データ |  emd_14134_validation.cif.gz emd_14134_validation.cif.gz | 10.4 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14134 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14134 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14134 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14134 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_14134.map.gz / 形式: CCP4 / 大きさ: 1000 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_14134.map.gz / 形式: CCP4 / 大きさ: 1000 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Composite of 3 globally sharpened focus-refined maps generated using Phenix Combine Focused Maps | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.7496 Å | ||||||||||||||||||||||||||||||||||||
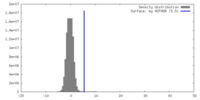
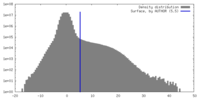
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
+全体 : Mitochondrial complex I in the deactive state with a ligand bound
+超分子 #1: Mitochondrial complex I in the deactive state with a ligand bound
+分子 #1: NADH-ubiquinone oxidoreductase chain 3
+分子 #2: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial
+分子 #3: NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial
+分子 #4: NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial
+分子 #5: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial
+分子 #6: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial
+分子 #7: NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial
+分子 #8: NADH-ubiquinone oxidoreductase chain 1
+分子 #9: NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial
+分子 #10: NADH-ubiquinone oxidoreductase chain 6
+分子 #11: NADH-ubiquinone oxidoreductase chain 4L
+分子 #12: NADH-ubiquinone oxidoreductase chain 5
+分子 #13: NADH-ubiquinone oxidoreductase chain 4
+分子 #14: NADH-ubiquinone oxidoreductase chain 2
+分子 #15: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mi...
+分子 #16: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mit...
+分子 #17: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial
+分子 #18: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial
+分子 #19: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2
+分子 #20: Acyl carrier protein, mitochondrial
+分子 #21: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5
+分子 #22: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6
+分子 #23: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8
+分子 #24: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11
+分子 #25: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13
+分子 #26: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1
+分子 #27: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 3
+分子 #28: NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial
+分子 #29: NADH dehydrogenase [ubiquinone] 1 subunit C2
+分子 #30: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5
+分子 #31: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1
+分子 #32: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mit...
+分子 #33: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mito...
+分子 #34: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 6
+分子 #35: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2, mito...
+分子 #36: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3
+分子 #37: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mito...
+分子 #38: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4
+分子 #39: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9
+分子 #40: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7
+分子 #41: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10
+分子 #42: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12
+分子 #43: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7
+分子 #44: NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial
+分子 #45: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+分子 #46: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+分子 #47: IRON/SULFUR CLUSTER
+分子 #48: FE2/S2 (INORGANIC) CLUSTER
+分子 #49: FLAVIN MONONUCLEOTIDE
+分子 #50: POTASSIUM ION
+分子 #51: GLYCEROL
+分子 #52: DODECYL-BETA-D-MALTOSIDE
+分子 #53: CARDIOLIPIN
+分子 #54: GUANOSINE-5'-TRIPHOSPHATE
+分子 #55: MAGNESIUM ION
+分子 #56: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
+分子 #57: ZINC ION
+分子 #58: ~{S}-[2-[3-[[(2~{R})-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butan...
+分子 #59: CHOLIC ACID
+分子 #60: MYRISTIC ACID
+分子 #61: water
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 4.78 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.5 構成要素:
| |||||||||
| グリッド | モデル: UltrAuFoil R0.6/1 / 材質: GOLD / 前処理 - タイプ: GLOW DISCHARGE 詳細: Following glow discharge for 90 s at 20 mA, the grid was treated for 48 hours in an anaerobic glovebox in ethanol containing 5 mM 11-mercaptoundecyl hexaethyleneglycol, washed three times in ...詳細: Following glow discharge for 90 s at 20 mA, the grid was treated for 48 hours in an anaerobic glovebox in ethanol containing 5 mM 11-mercaptoundecyl hexaethyleneglycol, washed three times in ethanol and dried prior to blotting | |||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 95 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK IV / 詳細: blot for 10 seconds before plunging. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) デジタル化 - サイズ - 横: 5760 pixel / デジタル化 - サイズ - 縦: 4092 pixel / 撮影したグリッド数: 2 / 実像数: 6780 / 平均露光時間: 2.4 sec. / 平均電子線量: 40.5 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 70.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.4 µm / 最小 デフォーカス(公称値): 1.0 µm / 倍率(公称値): 81000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー






























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)